Abstract
The membrane pore-forming activities of the antimicrobial peptide GWH1 have been evaluated in combination with the CXCR4-binding properties of the peptide T22, in self-assembling protein nanoparticles with high clinical potential. The resulting materials, of 25 nm in size and with regular morphologies, show a dramatically improved cell penetrability into CXCR4+ cells (more than 10-fold) and enhanced endosomal escape (the lysosomal degradation dropping from 90% to 50%), when compared with equivalent protein nanoparticles lacking GWH1. These data reveal that GWH1 retains its potent membrane activity in form of nanostructured protein complexes. On the other hand, the specificity of T22 in the CXCR4 receptor binding is subsequently minimized but, unexpectedly, not abolished by the presence of the antimicrobial peptide. The functional combination T22-GWH1 results in 30% of the nanoparticles entering cells via CXCR4 while also exploiting pore-based uptake. Such functional materials are capable to selectively deliver highly potent cytotoxic drugs upon chemical conjugation, promoting CXCR4-dependent cell death. These data support the further development of GWH1-empowered cell-targeted proteins as nanoscale drug carriers for precision medicines. This is a very promising approach to overcome lysosomal degradation of protein nanostructured materials with therapeutic value.
Export citation and abstract BibTeX RIS
Introduction
Cell-targeted drug delivery requires appropriate nanoscale vehicles (usually nanoparticles) for the generation of nanoconjugates [1]. These carriers have to be functionalized with appropriate ligands (usually peptides or proteins) that selectively bind to cell surface receptors overexpressed in target cells [2]. Selective delivery is specially desired in oncology, in which conventional therapy is majorly based on the systemic administration of untargeted chemotherapeutic drugs, associated to severe life-threatening side toxicity [3–7]. A nanoscale size of the conjugate, ranging from 10 to 100 nm, allows exploiting the enhanced permeability and retention (EPR) effect (based on the higher blood vessel permeability in tumoral tissues) while avoiding renal clearance (with a cut-off around 6–8 nm) and undesired aggregation in lung capillaries [8]. A diversity of materials is explored as nanoscale drug carriers including polymers, ceramics, metals and carbon nanotubes. The xenobiotic nature and potential toxicity of most of them pose severe concerns about their biosafety, at both individual and environmental levels [1, 9–18]. Contrarily, self-assembling proteins are promising, fully biocompatible nanostructured materials for drug delivery [19, 20]. The development of nanosized protein materials is strongly pusher by emerging nanobiotechnologies [20–24], that allow the engineering of proteins to confer self-assembling at the nanoscale. Being functionally versatile, proteins and protein materials can perform complex activities such as precise cell targeting, by the incorporation of peptide ligands in modular polypeptides, that act as building blocks of nanoscale entities [25]. In this context, T22 is a highly selective cationic ligand of the cytokine receptor CXCR4, that is overexpressed in about 20 human neoplasias and that correlates with aggressiveness and metastasis [26–32]. T22 promotes the endosomal-mediated internalization of non-amyloidal, self-assembling protein nanoparticles (formed by the T22-GFP-H6 fusion protein as building block), that result in the range size of 12 and 20 nm [33]. Selective T22-mediated binding and internalization result in a high level of specificity both in vitro and in vivo [34–36], and in an optimal biodistribution of the material in colorectal cancer animal models, when systemically administered [37]. In this context, T22-GFP-H6 nanoparticles have been recently used as potent nanocarriers of conventional antitumoral drugs [38], and closely related protein nanoparticles as vehicles for specific cytotoxic protein delivery in different cancer animal models [39, 40], both approaches with high therapeutic impact.
In protein-based nanoparticles, functional recruitment by protein fusion technologies [40–42] might be useful to enhance the penetrability of this construct (and similar nanoscale protein materials), which is now moderate, in order to improve its applicability as drug carrier. In this regard, poor endosomal escape and consequent lysosomal degradation may be a critical bottleneck for T22-GFP-H6 to efficiently deliver cytotoxic drugs into the cytoplasm, as generically observed for proteins and confirmed in our laboratory for CXCR4-targeted constructs (U. Unzueta, unpublished data). Since self-assembling proteins are highly versatile materials, we have here studied the combined activity of T22 and that of a potent pore-forming protein, the antimicrobial peptide (AMP) GWH1, simultaneously displayed on the surface of protein nanoparticles. Although highly promising in nanomedical applications, the combination of cell-targeting peptides and enhancers of cell penetrability has been so far poorly studied, while it might offer a way to overcome lysosomal degradation and limited functionality in the target cell. The data presented here demonstrate that GWH1 shows potent endosomolytic activity that increases transfection efficiently of functionalized protein nanoparticles into the cytoplasm. Despite this fact, the CXCR4 binding specificity is conserved, although reduced, in these materials. The functional combination provided by the peptide pair is then a promising approach to enhance the performance of smart drug nanocarriers based on functional peptides.
Materials and methods
Nanoparticle production and characterization
Self-assembling modular proteins T22-GFP-H6 [34], T22-GWH1-GFP-H6 [42] and GWH1-GFP-H6 [43] (figure 1(A)) have been described elsewhere. T22 is a powerful ligand of the cell surface cytokine receptor CXCR4 [34], overexpressed in more than 20 aggressive human neoplasias [28, 30, 44] and that acts as a highly convenient target for the precision delivery of antitumoral and antimetastatic drugs [38–40]. T22 mediates the endosomal internalization of fusion proteins that contain the peptide, in CXCR4+ cells, both in vitro and in vivo [34, 36]. These gene fusions were expressed from the plasmid pET22b in Escherichia coli Origami B (BL21, OmpT-, Lon-, TrxB-, Gor-, Novagen) under standard conditions [37]. Cells were disrupted in a French Press (Thermo FA-078A) at 1200 psi to obtain the soluble fraction. Protein purification was carried out through the His-tag by Immobilized Metal Affinity Chromatography (IMAC) using a HiTrap Chelating HP 1 ml column (GE Healthcare) with an AKTA purifier FPLC (GE Healthcare) [45]. Proteins were finally dialyzed against sodium bicarbonate buffer with salt (166 mM NaHCO3 pH 8 + 333 mM NaCl). Protein purity and integrity were checked by mass spectrometry (MALDI-TOF) and protein amounts by the Bradford assay.
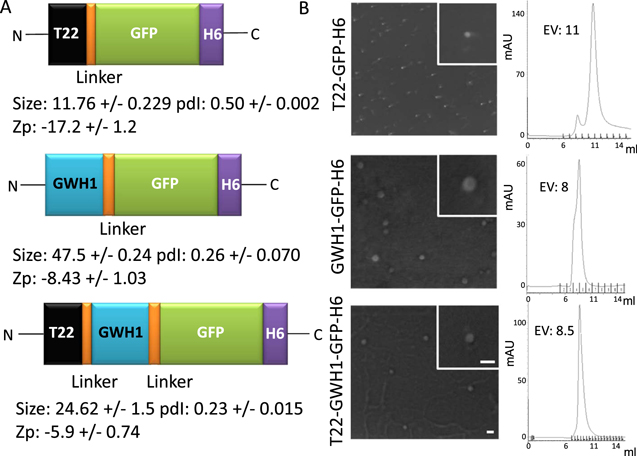
Figure 1. Features of GWH1-carrying nanoparticles. (A). Modular organization of the set of protein building blocks used in this study. All the shown polypeptides self-assemble as fluorescent nanoparticles. Hydrodynamic size (in nm, measured by DLS) and polydispersion index (pdI) are shown at the bottom of each cartoon and Z-potential (Zp) of the nanoparticles (in mV) are also indicated. (B). FESEM examination and size-exclusion chromatograms of purified nanoparticles monitored by UV detector at 280 nm. Bars indicate 50 nm, and all images have the same magnification. EV indicates the elution volume.
Download figure:
Standard image High-resolution imageMorphometric (size and shape) and ultrastructural characterization
The volume size distribution of nanoparticles was determined by dynamic light scattering (DLS) at 633 nm (Zetasizer Nano ZS, Malvern Instruments Limited, Malvern). The surface charge of proteins was assessed by zeta potential (Zp). Zp was measured at 25 °C and 633 nm, and DTS1070-folded capillary cell was used as sample container (Zetasizer Nano ZS, Malvern Instruments Limited). For quantitative analyses, three replicas of each sample were analyzed and the error in the measurements were estimated. Shape of nanoparticles was evaluated with a field emission scanning electron microscope (FESEM) Zeiss Merlin (Zeiss) equipped with a high resolution in-lens secondary electron detector and operating at 1 kV. To estimate the molecular mass of the purified nanoparticles size-exclusion chromatography (SEC) was performed on a Superdex 200 10/300 column (GE Healthcare). The column was previously calibrated with molecular mass standards (GE Healthcare) and injected protein nanoparticles (250 μl each) were eluted with a sodium bicarbonate buffer.
Membrane preparation and circular dichroism (CD)
Multilamellar vesicles were prepared by evaporating, under a stream of nitrogen, chloroform:methanol (2:1 v/v) from a solution of pure egg phosphatidyl choline (EPC) (Sigma-Aldrich). The dry lipid was resuspended in sodium bicarbonate buffer with salt at 1 mg ml−1 by repeating six consecutive cycles of heating for 2 min at 21 °C (a temperature above the phospholipid Tc) and vortexing for 1 min In these conditions, EPC self-aggregates into multilamellar vesicles (MLVs) [46]. Small unilamellar vesicles (SUVs) were prepared using a high intensity sonicator Branson sonifier 450, with 3 mm-diameter titanium probe. 1 ml of MLVs dispersion containing 4 mg ml−1 of EPC maintained on ice was sonicated for six cycles of 20 s, each one with 50% pulses (0.5 s on and 0.5 s off). The last sample was centrifuged at 15 000 g × 15 min to discard big aggregates and titanium particles.
Far-UV CD was measured at 25 °C in a Jasco J-715 spectropolarimeter to assess secondary structure information. The concentration of T22-GFP-H6 and T22-GWH1-GFP-H6 was adjusted to 0.2 mg ml−1 sodium bicarbonate buffer with salt. The protein spectrum was measured in the absence or the presence of small unillamellar vesicles (SUVs) (phosphatidylcholine at 0.5 and 1 mg ml−1). Samples were analyzed with a 1 mm path length cuvette. CD spectra were obtained over a wavelength range of 190–240 nm at a scan rate of 50 nm min−1 a response of 1 s and a bandwidth of 1 nm. Six scans were accumulated.
Cell culture and cell viability assay
Cervical and colorectal cancer cell lines were used to study the performance of recombinant proteins in vitro ( CXCR4+ HeLa, CXCR4+ SW1417 and CXCR4- SW1417 cells). HeLa cells were cultured in MEM Alpha (Minimum Essential Medium α, Gibco) supplemented with 10% fetal calf serum (Gibco) at 37 °C and 5% CO2 in a humidified atmosphere whereas SW1417 cells in Dulbecco's Modified Eagle's Medium (Gibco). To explore internalization and CXCR4 specific uptake of proteins, culture media was exchanged for serum-free Optipro medium (Gibco) supplemented with L-glutamine prior to the addition of nanoparticles. T22-GWH1-GFP-H6 and control T22-GFP-H6 and GWH1-GFP-H6 nanoparticles were added at 25 nM during 2 h in HeLa and SW1417 cells. Specific internalization through CXCR4 receptor was proved adding 1 h prior protein incubation a specific antagonist of CXCR4, AMD3100 at a ratio of 1:10. The CellTiter-Glo® Luminescent Cell Viability Assay (Promega) was used to determine the cytotoxicity of protein nanoparticles. For that, HeLa cells were plated in opaque-walled 96-well plates at 3500 cells/well in DMEM supplemented with 10% fetal calf serum for 24 h at 37 °C until reaching 70% confluence. Then, cells were incubated in presence of 2, 4, 8, 12 and 24 μM nanoparticles during 48 h at 37 °C. Subsequently, 100 μl of the single reagent (CellTiter-Glo® Reagent) was added directly to cultured cells and the plates were measured in the Multilabel Plater Reader VICTOR3 (PerkinElmer).
Internalization and endosomal degradation assays
To determine the threshold concentration for endosomal membrane destabilization, T22-GWH1-GFP-H6 and control T22-GFP-H6 and GWH1-GFP-H6 nanoparticles were added at 0.5, 1, 2, 3 and 4 μM and left for 24 h. To analyze endosomal escape of proteins, HeLa cells were incubated in absence and in presence of 100 μM chloroquine for 4 h before the addition of the protein at 1, 2, 3 and 4 μM. This drug is a lysosomotropic agent that induces vesicular rupture through protonation, in the acidic environment of late endosomes and lysosomes [47, 48]. Since chloroquine allows endosomal release and prevents lysosomal proteolysis [49], the increase in the intracellular material upon chloroquine addition is observed as indicative of the extent of lysosomal degradation of internalized protein in absence of the drug.
The uptake kinetics were recorded by exposing cells to nanoparticles at 1 and 4 μM for, 2, 5.5, 11, 16, 21 and 24 h prior to fluorescence measurement. Again, chloroquine was used for the determination of endosomal escape. Internalization was analyzed by detaching the cells with 1 mg ml−1 trypsin-EDTA (Gibco®) for 15 min, in a protocol specifically designed to remove externally attached protein [50]. The samples were analyzed by a FACS-Canto system (Becton Dickinson) using a 15 mW air-cooled argon ion laser at 488 nm excitation. All experiments were performed in duplicate. Fluorescence data recorded by cytometry were corrected by the specific fluorescence of the protein to render comparative units in terms of protein amount. The specific fluorescence was previously determined by a Varian Cary Eclipse fluorescence spectrophotometer (Agilent Technologies) at 510 nm using an excitation wavelength of 488 nm.
Confocal laser scanning microscopy
For confocal microscopy, HeLa cells were grown on Mat-Tek plates (MatTek Corporation). Upon exposure to nanoparticles at 4 μM for 24 h, cell nuclei were labeled with 5 μg ml−1 Hoechst 33342 (ThermoFischer) and the plasma membrane with 2.5 μg ml−1 CellMaskTM Deep Red (ThermoFischer) for 10 min at room temperature. Cells were then washed in PBS (Sigma-Aldrich Chemie GmbH). Live cells were recorded with a TCS-SP5 confocal laser scanning microscope (Leica Microsystems) using 63x (1.4 NA) oil immersion objective lenses. Hoechst 33 342 labeled DNA was excited with a blue diode (405 nm) and detected in the 415–460 nm range. GFP-proteins were excited with an Ar laser (488 nm) and detected in the 525–545 nm range. CellMask was excited with a HeNe laser (633 nm) and detected in the 650–775 nm range. The confocal pinhole was set to 1 Airy unit and z-stacks acquisition intervals were selected to satisfy Nyquist sampling criteria. Three-dimensional images were obtained using the Surpass Module in Imaris X64 v.7.2.1. software (Bitplane).
Synthesis and characterization of the T22-GWH1-GFP-H6-Auristatin nanoconjugate
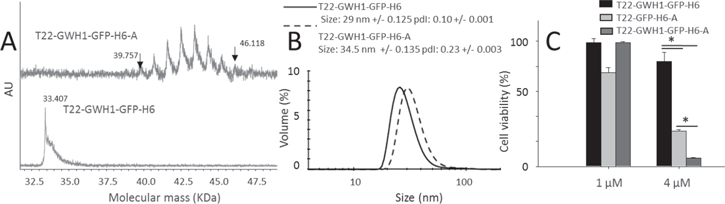
The T22-GWH1-GFP-H6-Auristatin nanoconjugate (T22-GWH1-GFP-H6-A) was synthesized by covalent binding of the protein nanoparticle and Auristatin. The final conjugate was obtained by reacting maleimide groups of Auristatin with primary amines of nanoparticles (like lysine and arginine sidechains) favored at pH = 8. The quality analysis of the conjugation was performed by MALDI-TOF spectra. The molecular mass of the T22-GWH1-GFP-H6-A carrying from 7 to 14 auristatin molecules (911 Da each) was identified by mass spectrometry. Next, we studied T22-GWH1-GFP-H6-A cytotoxic activity measuring cell viability and using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega), following manufacturer recommendations. To that purpose, we exposed CXCR4+ HeLa cells to T22-GWH1-GFP-H6-A at 1 and 4 μM for 24 h as compared to equimolecular concentrations of T22-GFP-H6-A or T22-GWH1-GFP-H6.
Statistics
All the experiments were performed in triplicate. One-way ANOVA followed by Fisher's least significant difference (LSD) method was used for multiple comparisons. Pairwise comparisons were performed using Student-t tests. Statistical differences (indicated as * in the Figures) were assumed at p < 0.01. Microsoft Excel was used for all statistical analyses.
Results
T22-GWH1-GFP-H6 (figure 1(A)) contains the AMP GWH1 inserted as an additional module between the N-terminal CXCR4 ligand T22, and the core GFP. This protein, as well as the related constructs T22-GFP-H6 and GWH1-GFP-H6 (figure 1(A)), form regular nanoparticles with a toroid-like morphology, because of the cationic character of the N-terminal region [51]. These materials show distinguishable Z-potential values and nanoscale sizes (figure 1(A), (B)). In particular, T22-GWH1-GFP-H6 organizes as structures sizing 25 nm in average, being larger than the parental T22-GFP-H6 (12 nm) and smaller than the shorter GWH1-GFP-H6 (47 nm). SEC analysis of the three materials revealed different elution peaks, from which we estimated the structural composition of the particles in 31, 11 and 45 monomers respectively. The value for T22-GFP-H6 (formed by 11 building blocks), fits reasonably well with a previous in silico modeling of the oligomeric nanoparticle, that predicted the material being composed by 10 regularly accommodated GFP monomers [37].
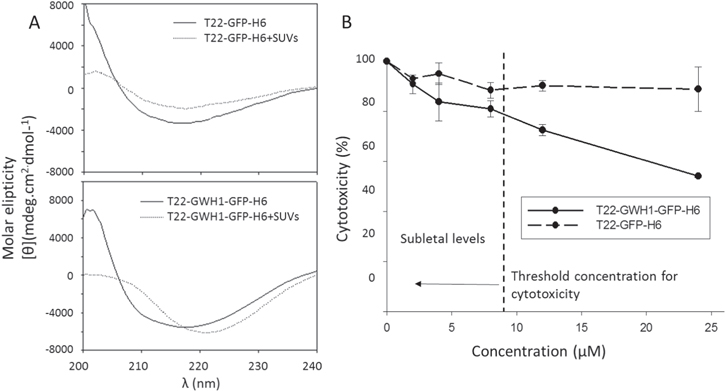
We were interested in knowing if GWH1, a pore-forming peptide, might enhance the penetrability of the protein construct mediated by the specific interaction between T22 and CXCR4, upon endosomal membrane destabilization. GWH1 exerts its cytolytic activity by folding into an amphipathic helix upon selective binding and insertion into the target membrane. In order to investigate whether the addition of the T22 moiety and the nanoparticulated form itself might affect the GWH1 ability to form α-helical structures, we measured the peptide conformation by circular dichroism (CD) in the absence or the presence of small unillamellar vesicles (SUVs). The T22-GWH1-GFP-H6-spectrum in the absence of SUVs shows the typical behavior of a β-strand pattern corresponding to the expected beta-barrel structure of the GFP (figure 2(A)), with a minimum around 218 nm [52]. By contrast, when these nanoparticles interact with SUVs a qualitative change is observed in the DC spectrum in which the minimum moved to 222 nm (figure 2(A)) corresponding to the appearance of novel helical conformation. Interestingly T22-GFP-H6 did not exhibit this qualitative change in CD spectrum as it interacted with SUVs (figure 2(A), bottom).
Figure 2. Interaction of GWH1-containing nanoparticles with cell membranes. (A). T22-GWH1-GFP-H6 and T22-GFP-H6 CD spectra at wavelength range of 200–240 nm. In the absence of SUVs, both spectra of proteins indicate the occurrence of β-sheet conformation. The CD spectra of T22-GWH1-GFP-H6, in presence of membranes, shows the appearance of the helical content. (B). Intrinsic cytotoxicity on HeLa cells imposed by the AMP GWH1. Cells were incubated in presence of 2, 4, 8, 12 and 24 μM of protein assembled as nanoparticles during 48 h. T22-GFP-H6 does not show any biological effect at these doses. A threshold for GWH1 toxicity is shown.
Download figure:
Standard image High-resolution imageAMPs, as effective self-defense tools, exhibit a threshold concentration (called the lethal concentration) for their membrane activity on eukaryotic cells, below which no effect is observed [53]. This security level allows the antimicrobial activity at low concentrations, without harming own body cells. Since it had been previously described that GWH1 exhibits cytotoxic effects over cancer cells with a reported threshold ranging from 20 to 250 μM, we first determined the intrinsic cytotoxicity of T22-GWH1-GFP-H6 over CXCR4+ HeLa cells. As observed (figure 2(B)), T22-GWH1-GFP-H6 nanoparticles showed a dose-dependent cytotoxicity with a significant lethal concentration lower than the predicted for the GWH1 peptide alone. This cell killing effect, when properly targeted to tumoral tissues, has proved to be exploitable to design antitumoral drugs [42]. Under 8 μM, no cytotoxicity was observed in HeLa cells upon exposure to T22-GWH1-GFP-H6 nanoparticles, and further analysis of cell penetrability and specificity were performed in a safe margin, up to 4 μM.
Then, endosomal escape promoted by GWH1 was evaluated by comparison between T22-GWH1-GFP-H6 and its parental T22-GFP-H6. As observed (figure 3(A)), over 1.5 μM, T22-GWH1-GFP-H6 is dramatically more efficient than T22-GFP-H6 in escaping the endosome, suggesting a threshold concentration for endosomal membrane destabilization. This action cannot be performed by the control nanoparticle GWH1-GFP-H6, which lacks the cell surface ligand (T22) for internalization. Such endosomal escape protects the 40% of the internalized protein nanoparticles from lysosomal degradation (figure 3(B)), that is responsible for the destruction of most T22-GFP-H6. Interestingly, the presence of GWH1 impairs, contrarily, the CXCR4-dependence in the uptake of the protein materials (figure 3(C)) but it does not abort it. Less than 2% of T22-GFP-H6 enters target cells in the presence of the CXCR4 antagonist AMD3100, while almost 70% of T22-GWH1-GFP-H6 penetrates HeLa cells under the same conditions (figure 3(C)).
Figure 3. Cell penetration and endosomal escape of GWH1-containing nanoparticles. (A). Cell internalization of GWH1-empowered nanoparticles measured through intracellular fluorescence, after a harsh trypsin treatment to remove external material. A threshold for the endosomal escape properties is shown at 1.5 μM. FU are fluorescence units. (B). Fraction of internalized protein degraded in the endosomes, as measured by chloroquine addition. Cells were incubated in the absence and in the presence of 100 μM chloroquine for 4 h before the addition of the protein at 0.5, 1, 2, 3 and 4 μM during 24 h. (C). Specificity of CXCR4-mediated internalization of nanoparticles added at 25 nM visualized through the fraction of protein internalization inhibited by the CXCR4 antagonist, AMD3100 [54–56], incubated 1 h prior to protein treatment at a ratio of 1:10. * indicates significant differences between T22-GWH1-GFP-H6 data and that of the other proteins.
Download figure:
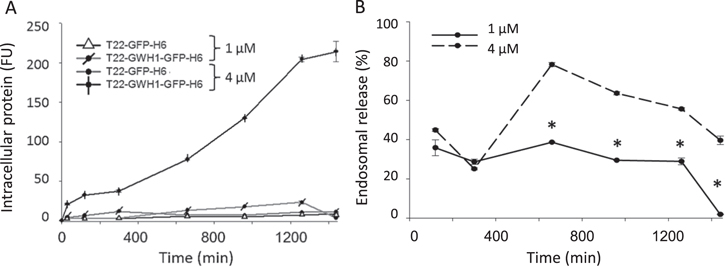
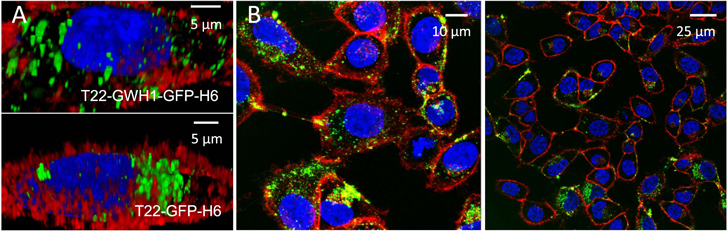
Standard image High-resolution imageIn accordance to the obtained results, at a concentration (1 μM) in which GWH1 does not promote endosomal escape, the cell uptake of T22-GWH1-GFP-H6 is slightly higher than that of the parental T22-GFP-H6 (figure 4(A)). The uptake slows down at about 5 h, reaching a steady constant intracellular concentration during 24 h. At a concentration above that threshold (4 μM), however, the penetration of T22-GWH1-GFP-H6 is extremely efficient. The amount of intracellular protein keeps increasing, without reaching any plateau at least during 24 h of exposure. During this experiment time, T22-GWH1-GFP-H6 is majorly degraded at 1 μM, but only 60% of the material is destroyed at 24 h in the endosomes at 4 μM (figure 4(B)). This observation confirms again, the endosomal escape of the nanoparticles promoted by GWH1. Under this situation, most of T22-GWH1-GFP-H6 is found homogenously distributed by the cell cytoplasm, while the parental T22-GFP-H6 is majorly concentrated in a perinuclear region, as dissected by 3D confocal reconstructions (figure 5(A)). The perinuclear localization of T22-GFP-H6 fits with previous data that suggested a strong endosomal retention of this material [34]. The broadest intracellular distribution of T22-GWH1-GFP-H6 was confirmed by wide 2D confocal imaging of larger areas of cultured cells, at different magnifications (figure 5(B)).
Figure 4. Dynamics of cell penetration of GWH1-containing nanoparticles. (A). Kinetics of cell penetration of GWH1-empowered nanoparticles. Relative amounts of intracellular nanoparticles penetrating HeLa cells at different times after exposure (at 2 different concentrations). (B). Endosomal release of T22-GWH1-GFP-H6 under the same conditions. * indicates significant differences between T22-GWH1-GFP-H6 data and that of the other proteins.
Download figure:
Standard image High-resolution imageFigure 5. Intracellular localization and fate of protein nanoparticles. (A). 3D confocal reconstructions of cultured HeLa cells exposed to 4 μM of GWH1-empowered nanoparticles and the control T22-GFP-H6. Blue label corresponds to the nucleus, red label to membranes and green label is the natural fluorescence of the nanoparticles. (B). Broad confocal fields, at different magnifications, showing the intracellular localization of T22-GWH1-GFP-H6 homogenously distributed by the cell cytoplasm. All images were taken 24 h after exposure.
Download figure:
Standard image High-resolution imageThe high cell penetrability and improved endosomal escape conferred by the AMP to the protein nanoparticles, combined with the significant level of CXCR4 selectivity (figures 3, 4 and 5) prompted us to explore its potentiality as vehicles for targeted drug delivery, and therefore their potentiality for clinical applications. For that, we chemically coupled the antitumoral drug Auristatin to T22-GWH1-GFP-H6, and explored the ability of the nanoconjugate (T22-GWH1-GFP-H6-A) to deliver the drug in the cell cytoplasm. Auristatin is a potent antimitotic peptide derived from the mollusc Dolabella auricularia, with applicability in targeted therapies in a spectrum of tumors [57]. The chemical conjugation rendered a narrow spectrum of nanoconjugates ranging from 7 to 14 drug molecules per nanoparticle, as determined by MALDI (figure 6(A)), and it slightly modified the volume and Zp of the materials (figure 6(B)). When CXCR4+ cells were exposed to drug-loaded nanoparticles, a dramatic impact on cell viability was observed (figure 6(C)). Importantly, the nanoparticle version lacking the AMP (T22-GFP-H6-A) was less efficient in promoting cell death (figure 6(C)), confirming again the positive impact of this peptide in the release of the whole complex, including the drug, to the cytoplasm.
Figure 6. Characterization of T22-GWH1-GFP-H6-Auristatin nanoconjugates. (A). Mass spectrometry of T22-GWH1-GFP-H6-Auristatin (labeled as A) upon chemical conjugation. Eight peaks were identified corresponding to nanoparticles with seven to fourteen attached drug molecules. Flanking peaks are indicated with arrows, and also their molecular masses. The non-labeled peaks correspond to molecular masses of 40.682, 41.601, 42.500, 43.413, 44.320 and 45.244 kDa respectively. At the bottom, the spectrum of drug-free nanoparticles. (B). Hydrodynamic size distribution of T22-GWH1-GFP-H6-A nanoconjugate compared to the drug-free nanoparticle. (C). Reduction in cell viability of HeLa cells exposed to T22-GWH1-GFP-H6-A for 48 h, as compared to T22-GFP-H6-A and T22-GWH1-GFP-H6. * indicates significant differences between T22-GWH1-GFP-H6-A data and that of the other proteins.
Download figure:
Standard image High-resolution imageDiscussion
Protein-based drugs are of high interest in molecular medicines as they can be produced among a spectrum of cell factories by simple recombinant DNA technologies [58]. In cancer therapies, modular approaches based on fusion protein engineering [41] allow recruiting, in single chain polypeptides, diverse functions required for efficient cell targeting, internalization and endosomal escape, that are encoded by proteins or protein domains [25, 59]. Self-assembling, that can be provided by short peptide stretches [20], allows the proteins being presented as regular oligomers within the nanoscale size, a presentation that enhances their tumoral accumulation by physical mechanisms linked to the EPR [8]. In a step further beyond the simple protein-drug association principles (on which Abraxane® is based [60]), nanostructured, cell targeted proteins can be ideal vehicles for drug targeting in cancer therapy in form of drug nanoconjugates [3]. We have here tested the combination of the AMP GWH1 [61], a potent membrane pore former, with T22 [34], a potent CXCR4 ligand, in protein-only CXCR4-targeted nanoparticles, regarding efficiency and specificity of CXCR4+ cell binding and penetrability. GWH1 is a synthetic AMP designed to show enhanced antimicrobial activity while reduced cell killing activity on normal eukaryotic cells, such as 3T3 fibroblasts [62] or erythrocytes (with low haemolytic activity) [63]. It also shows improved selectivity for surface binding and killing of cancer cells as compared to normal cells, because of similarly to bacteria, their membrane is enriched in anionic components [64]. GWH1 exerts its cytolytic activity by folding into an amphipathic helix upon selectively binding and insertion into the target membrane, leading to breakdown of the membrane structure, thus causing leakage of cell contents, finally resulting in cell death. Being used as a synthetic peptide alone, GWH1 is fully functional in form of fusion proteins [43], what opens the door to consider its inclusion in multifunctional constructs. The combination of pore-forming and cell-targeting agents, despite their obvious interest in cancer therapies, has not been systematically explored.
In this context, we have demonstrated here that the accommodation of GWH1 in longer modular polypeptides containing the cationic CXCR4 ligand T22 (figure 1(A)) does not affect neither the functionality of GWH1 nor the ability of the T22- and H6-empowered proteins to form regular nanoparticles (figure 1(A)). Moreover, a clear membrane-disrupting ability of T22-GWH1-GFP-H6 was shown over cancer cells with the expected threshold concentration (figure 2), indicating the ability of GWH1 to create pores even in a nanoparticulated version. This fact can be obviously exploited to construct therapeutic protein materials based on functional recruitment, through simple fusion technologies. Interestingly, at sub-lethal concentrations, the GWH1 module enhances rapid and effective translocation of T22-GWH1-GFP-H6 nanoparticles from the endocytic vesicles into cytoplasm, when comparing with the parenteral T22-GFP-H6 (figure 3). Also, despite its observable endosomolytic activity, GWH1 does not promote release from vesicles into the cytoplasm except when administered at concentrations above 1.5 μM. This behavior is compatible with molecular models currently proposed to explain the permeability properties of α-helical antimicrobial sequences and the need of a threshold concentration for pore formation [53, 65, 66]. In this context, a punctuate fluorescence pattern was observed for internalized T22-GFP-H6 (figure 5), indicating endosome permanence. In contrast, a different intracellular fluorescence pattern was shown by T22-GWH1-GFP-H6, characterized by a more homogeneous distribution of the material in the cytoplasm, consistent with the lysosomal release of a significant fraction of the recombinant cargo (figure 5).
Interestingly, membrane pore formation, although a fully unspecific and very efficient process, does not abort the specific CXCR4-dependent cell penetration mediated by T22. CXCR4 specificity is however reduced by the presence of GWH1 in the protein constructs from more than 90% to 30% (figure 3(C)). This indicates that enhanced cell penetrability though pore formation cannot be gained simultaneously to a high cell specificity, as previously observed when exploring the fusogenic peptide HA2 from the influenza virus hemagglutinin [67]. However, since the in vivo tumor targeting of antibody-drug conjugates has determined to be around 1% [1, 18], 30% of receptor specificity appears as still good data. In this context, the protein nanoparticles generated here and empowered with GWH1 have been efficiently loaded with the antitumoral drug Auristatin by chemical conjugation (figure 6). The presence of the AMP in the material dramatically improved the cytotoxicity of the drug in the nanoconjugate when exposed to cultured CXCR4+ cell (figure 6(C)), when compared to the use of the parental T22-GFP-H6 as vehicle. These data, combined with the confocal microscopy imaging (figure 5) and the experimental determining endosomal escape (figure 3) demonstrate the endosomolytic properties of GWH1 when integrated, as a functional domain, in modular self-assembling proteins. Obviously, the clinical impact of pore formation combined with tumor cell targeting should be explored further. Since T22-based protein nanoconstructs have been recently shown extremely powerful for the precision delivery of chemical and protein antitumoral drugs [38–40, 42], the improvement of their intracellular trafficking properties, exploiting the functional versatility of protein materials, would be extremely desirable.
Conclusion
The obtained set of data stresses the strong value of membrane active peptides used as potential enhancers of drug penetrability and endosomal escape into target cells, when displayed in nanoscale supramolecular complexes. This has been proved here by the incorporation of GWH1 to a cell-targeted nanoscale protein vehicle through conventional protein engineering, generating a stable and functional nanomaterial. In addition, the presented results also point out that membrane pore formation does not preclude specific cell targeting mediated by a partner driver peptide, although the receptor specificity is reduced. In the current context of innovative drug design and the generation of smart nanoscale vehicles as delivery agents, the data presented in this study offer critical clues regarding the recruitment of appropriate functional agents based on recombinant proteins. The successful incorporation of membrane pore forming peptides into self-assembling functional proteins offers a powerful approach to overcome one of the major bottlenecks in the endosomal delivery of protein materials, namely lysosomal degradation.
Acknowledgments
This study has been funded by the Agencia Estatal de Investigación (AEI) and Fondo Europeo de Desarrollo Regional (FEDER) (grant BIO2016-76063-R, AEI/FEDER, UE), AGAUR (2017SGR-229) and CIBER-BBN (project NANOPROTHER) granted to AV, Marató de TV3 foundation (TV32013-3930) and ISCIII (PI15/00272 co-founding FEDER) to EV and ISCIII (PI15/00378 and PIE15/00028, co-founding FEDER), Marató de TV3 foundation (TV32013-2030) and AGAUR 2014-PROD0005 to RM. Protein production has been partially performed by the ICTS 'NANBIOSIS', more specifically by the Protein Production Platform of CIBER-BBN/ IBB (http://www.nanbiosis.es/unit/u1-protein-production-platform-ppp/). We are indebted to SCAC (UAB) for cell culture facilities and assistance and to the Microscopy Service at the UAB. NS was supported by a predoctoral fellowship from the Government of Navarra, LSG was supported by AGAUR (2018FI_B2_00051), UU is supported by PERIS program from the health department of la Generalitat de Catalunya and JMS is a Career Investigator from CONICET. AV received an ICREA ACADEMIA award.