Abstract
Cryoconite is a microbe–mineral aggregate which darkens the ice surface of glaciers. Microbial process and marker gene PCR-dependent measurements reveal active and diverse cryoconite microbial communities on polar glaciers. Here, we provide the first report of a cryoconite metagenome and culture-independent study of alpine cryoconite microbial diversity. We assembled 1.2 Gbp of metagenomic DNA sequenced using an Illumina HiScanSQ from cryoconite holes across the ablation zone of Rotmoosferner in the Austrian Alps. The metagenome revealed a bacterially-dominated community, with Proteobacteria (62% of bacterial-assigned contigs) and Bacteroidetes (14%) considerably more abundant than Cyanobacteria (2.5%). Streptophyte DNA dominated the eukaryotic metagenome. Functional genes linked to N, Fe, S and P cycling illustrated an acquisitive trend and a nitrogen cycle based upon efficient ammonia recycling. A comparison of 32 metagenome datasets revealed a similarity in functional profiles between the cryoconite and metagenomes characterized from other cold microbe–mineral aggregates. Overall, the metagenomic snapshot reveals the cryoconite ecosystem of this alpine glacier as dependent on scavenging carbon and nutrients from allochthonous sources, in particular mosses transported by wind from ice-marginal habitats, consistent with net heterotrophy indicated by productivity measurements. A transition from singular snapshots of cryoconite metagenomes to comparative analyses is advocated.
Export citation and abstract BibTeX RIS

Content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
Cryoconite holes are thought to represent 'ice-cold hot-spots of microbial diversity and activity' (Edwards et al 2012) at the surface of Earth's largest freshwater biome, namely glaciers and ice-sheets (Hodson et al 2008, Shiklomanov 1993).
Consequently, these melt-water filled depressions which form by localized reduction of albedo by dark microbe–mineral aggregates known as cryoconite (Takeuchi 2002, Takeuchi et al 2001, Wharton et al 1985) have garnered growing interest. Investigations of microbial processes associated with cryoconite have ascribed global significance to the rates of supraglacial carbon cycling measured (Anesio et al 2009), and analysis of bacterial carbon fluxes reveals net accumulation of carbon by cryoconite (Anesio et al 2010). Furthermore, the reduction of albedo by cryoconite (Takeuchi 2002, Takeuchi et al 2001) along with other supraglacial microbial consortia (Yallop et al 2012) may contribute to the 'biological darkening' (Irvine-Fynn et al 2012) of glacier surfaces.
Therefore, better insight into the molecular foundations of cryoconite ecosystem functionality is sought. Building from a clone library from Antarctic cryoconite (Christner et al 2003), investigators have employed PCR-dependent methods to characterize cryoconite diversity. T-RFLP reveals distinct community structures for bacteria and fungi between neighbouring High Arctic glaciers and ice-marginal habitats (Edwards et al 2011, 2012, 2013), and between Arctic and Antarctic glaciers (Cameron et al 2012b), while clone libraries reveal communities dominated by either Proteobacteria, Cyanobacteria or both phyla (Cameron et al 2012b, Edwards et al 2011). Functional gene PCR detects nifH genes (Telling et al 2012) consistent with nitrogen fixation (Telling et al 2011, 2012), and other genes associated with carbon and nitrogen cycling (Cameron et al 2012a). Although the bacterial community structure of cryoconite is closely correlated with rates of primary production and respiration (Edwards et al 2011), further insights into community structure-function relationships are limited.
Moreover, previous studies focus on the cryoconite ecology of polar glaciers and ice-sheets. Less is known about the microbial diversity of cryoconite on alpine glaciers. Although the relative accessibility, vulnerability and economic importance (Kaltenborn et al 2010) of alpine glaciers provide an impetus for their study, particularly as cryoconite may accelerate the melting of alpine glaciers (Oerlemans et al 2009), culture-independent studies of alpine glacier biodiversity are limited to ice and melt-water (Franzetti et al 2013, Simon et al 2009, Wilhelm et al 2013).
The rapid democratization of high-throughput sequencing enabled metagenomics (Thomas et al 2012) provides an attractive means of gaining deeper insight into cryoconite ecosystem structure and functionality. In this study, we exploited the development of the Institute of Biological, Environmental and Rural Sciences (IBERS) Aberystwyth Illumina HiScanSQ Facility to provide a snapshot of the phylogenetic and functional diversity of an alpine cryoconite metagenome. To our knowledge, the present study is the first to describe a cryoconite metagenome, and the first application of Illumina sequencing platforms to gain insights into a glacial metagenome at sequencing depth much greater than previously possible with pyrosequencing (Simon et al 2009).
2. Materials and methods
2.1. Study site and sampling
Rotmoosferner is a north-facing valley glacier in the Ötztal Alps in Tirol, Austria located at 46°50'N, 11°03E presently terminating at approximately 2450 m asl, and designated an UNESCO Man-and-Biosphere Reserve. Rotmoosferner was previously partially fed by a second glacier, Wasserfallferner, but the rapid retreat and thinning of these glaciers (Abermann et al 2009) disconnected the glaciers in 2005. Rotmoosferner is shaded from afternoon sun by the ridgeline of Wasserfallferner (figure 1). Mean annual air temperature and precipitation are −1.3 ° C and 820 mm w.e. respectively, while the bedrock is dominated by amphibolite quartzo-feldspathic rocks (Kaufmann 2001). The >2 km retreat of Rotmoosferner has been documented since 1872, and the pace of retreat has increased this century (Anesio et al 2010). Consequently, the processes of foreland colonization and succession by plants, invertebrates and microbes over the Rotmoosferner foreland chronosequence has been studied (e.g. Erschbamer et al 2008, Kaufmann 2001, Philippot et al 2011) to a greater extent than its supraglacial ecology, although the bacterial secondary production and phylum level distribution of supraglacial debris have been reported (Anesio et al 2010, Philippot et al 2011).
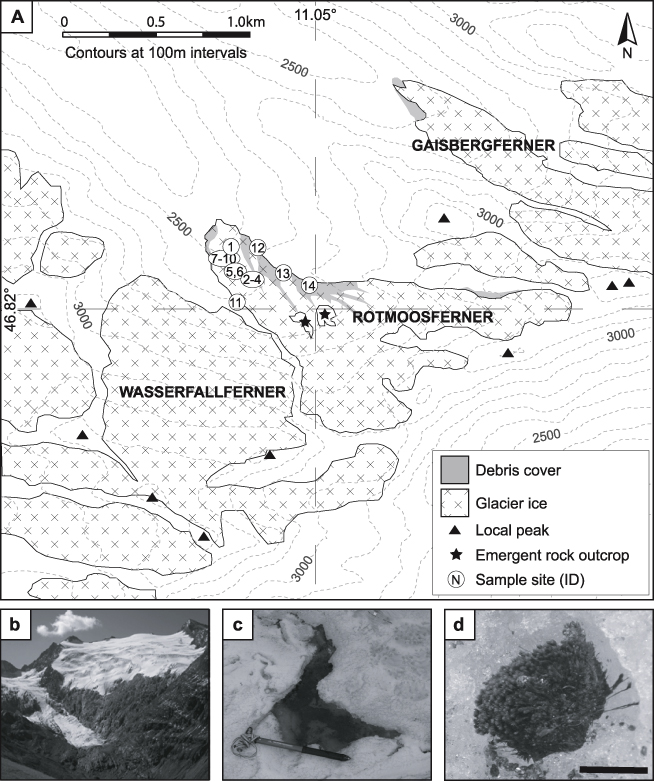
Figure 1. (A) Study location with cryoconite holes numbered. (B) Rotmoosferner's position beneath Wasserfallferner, (C) a typical cryoconite hole (R10) on Rotmoosferner (ice axe for scale, 50 cm shaft) and (D) a supraglacial moss associated with arthropods (scale approximately 3 cm).
Download figure:
Standard image High-resolution imageCryoconite was sampled on 8 and 14 September 2010 (two cryoconite holes on the first day, the remaining 12 on the second day) from 14 cryoconite holes in the ablation zone of Rotmoosferner. Debris were aspirated aseptically using plastic Pasteur pipettes into 15 ml centrifuge tubes and transported on ice to Alpine Forschungsstelle-Obergurgl within four hours, before freezing at −20 ° C for transfer to the Aberystwyth laboratory frozen in insulated containers. Additionally, in situ measurements of cryoconite bacterial primary and secondary production and estimation of net ecosystem production were conducted essentially as described by Anesio et al (2009) and Hodson et al (2010) respectively and detailed in the supplementary methods (available at stacks.iop.org/ERL/8/035003/mmedia).
2.2. Metagenome sequencing
Cryoconite community genomic DNA was extracted using Powersoil DNA kits (MoBio, Inc. Solana, California) as described in the supplementary methods. Where possible, equimolar quantities (214.3 ng per sample, except for Samples 3 (97 ng); 4 (15.5 ng), 7 (188 ng); 9 (151 ng); 14 (163.84 ng)) as measured by Quant-iT high sensitivity DNA assay, (Invitrogen, Inc. Paisley, Scotland) of DNA extracts were pooled for library preparation. Subsequently, the pooled DNA was sheared by 18 cycles of sonication (30 s cycles) using a Bioruptor (Diagenode, S.A., Liège, Belgium), and purified with a Qiagen PCR purification kit (Qiagen, Ltd Crawley, UK). The DNA was used to create an Illumina paired-end library with an average insert size of 360 base pairs (bp) according to the manufacturer's instructions (Paired-End Sample Prep Kit, Rev. E, Illumina, Ltd, Essex, UK), with the exception of 14 PCR cycles during the enrichment step. The library was sequenced at 2 × 51 bp using an Illumina HiScanSQ at the IBERS Aberystwyth Translational Genomics Facility according to standard procedures.
2.3. Data processing and analysis
The 535 million reads were optimally assembled using the De Brujin graph assembler AbySS 1.3.3 (Simpson et al 2009) and the optimal assemblies were performed with k-mer length of 31. The total assembly size was 1190 Mbp, in 9.7 million contigs, of average size 122 bp and N50 of 146 bp, of which the longest contig was 237 Kbp in size. Over 200 000 contigs were longer than 500 bp: their combined length was 366 Mbp.
The assembled metagenome was imported as a FASTA file into MG-RAST 3.2 (Meyer et al 2008) and annotated by comparison to the M5NR database (Wilke et al 2012). Investigations of phylogenetic diversity applied cutoffs of 60% minimum identity over 15 residues with an e value of 1 × 10−5 or less, while functionality comparisons utilized cutoff e value of 1 × 10−2 or less.
For functionality comparisons, the metagenome was compared to those from soil (e.g. Fierer et al 2012), biofilm-dominated matrices such as microbial mats (e.g. Varin et al 2012), sludge (Martin et al 2006) and aquatic habitats (NCBI project 33179 and (Khodadad and Foster 2012, Rusch et al 2007)) made using a variety of sequencing platforms and publically available on MG-RAST (see supplementary table 1 for details, available at stacks.iop.org/ERL/8/035003/mmedia). Read relative abundance per Subsystems (Overbeek et al 2005) top level functional category (at an e value of 1 × 10−2 or less) was used for Principal Components Analysis (PCA) and fourth-root Bray–Curtis similarities used for cluster analysis using PRIMER6 and PERMANOVA+ (version 1.0.2 and version 6.1.12; Primer-E, Ivybridge, UK). The only other glacial metagenome datasets publically available presently, composed of 454 pyrosequencing of glacial ice from a German glacier (Schneeferner, EBI-SRA SRA001163 Simon et al 2009), were ported into MG-RAST for comparison. The alpine cryoconite metagenome assembled in this study has been deposited on MG-RAST as 4 491 734.3.
3. Results and discussion
Ratios between in situ heterotrophic and autotrophic production rates of cryoconite sediments were calculated from the 12 cryoconite holes sampled on 14 September 2010. Primary production ranged from 2.56 × 10−2–19.8 μg C g−1 h−1, whereas secondary production ranged between 0.307–3.03 × 102 μg C g−1 h−1 (table 1). Except for two samples (R7 and R10), the majority showed a clear heterotrophic production rate. Furthermore, results for three light and dark batch incubations revealed net heterotrophy in the two cryoconite holes (R11 and R12) subjected to net ecosystem productivity estimation by change in dissolved inorganic carbon. The photosynthesis estimates (means of triplicate incubations of sediment from each hole = 9 and 29 μg C g−1 d−1 respectively) were lower than the respiration rates (mean 31 and 36 μg C g−1 d−1). In summary, the parallel in these observations support the contention of net heterotrophy in cryoconite at the time of sampling (Telling et al 2010), and demonstrate rates of microbial activities in Rotmoosferner cryoconite comparable to or greater than polar cryoconites (Anesio et al 2009).
Table 1. Radiometric measurement of primary (PP) and secondary production (SP) in Rotmoosferner cryoconites sampled for metagenome sequencing on 14 September 2010. Values are the mean of triplicate incubations, and cryoconites with a positive ratio of SP:PP consistent with net heterotrophy are highlighted in bold.
| Primary (PP) and secondary production (SP) in rotmoosferner cryoconite | |||
|---|---|---|---|
| Cryoconite hole | SP (μg C g−1 h−1) | PP (μg C g−1 h−1) | SP:PP ratio |
| R3 | 303.18 | 10.10 | 30.02 |
| R4 | 182.77 | 5.60 | 32.64 |
| R5 | 62.70 | 0.27 | 233.95 |
| R6 | 43.76 | 0.03 | 1709.31 |
| R7 | 0.31 | 5.16 | 0.06 |
| R8 | 83.79 | 0.11 | 775.85 |
| R9 | 66.06 | 1.74 | 37.97 |
| R10 | 0.28 | 19.80 | 0.01 |
| R11 | 47.41 | 0.08 | 585.33 |
| R12 | 173.75 | 0.40 | 431.14 |
| R13 | 23.90 | 0.60 | 39.53 |
| R14 | 51.09 | 0.70 | 72.60 |
Metagenomic DNA was successfully extracted from all 14 cryoconite debris samples collected from the ablation zone of Rotmoosferner in September 2010 (figures 1(A)–(D)). Comparison of the Rotmoosferner cryoconite ecosystem by pyrosequencing and T-RFLP to that of other glaciers will be reported elsewhere. The fourteen samples comprised all the cryoconite holes which could be found across the ablation zone at the time of sampling, but other cryoconite holes in proximity to crevasses may have been present.
As this study's objective was to provide a first metagenomic insight into the cryoconite ecosystem of an alpine glacier, and the yield of DNA per sample was limited, DNA samples from separate cryoconites were pooled for library production.
It is known that glacier-specific factors, for example geometry, surface hydrology and presumably lithology (Edwards et al 2011, 2012), can affect cryoconite microbial communities and that temporal variation is likely (Anesio and Laybourn-Parry 2012). Therefore, although our study design accounts for intra-glacier variation, our findings are subject to the caveat that the metagenome presented is probably most representative of cryoconite ecosystem genetic and functional diversity during the late growing season at Rotmoosferner, and that further insights are extrapolative in nature.
The optimal assembly comprised 9 727 829 sequences totalling 1.19 × 109 bp with a mean sequence length of 122 bp (±534 bp, ±1SD). Over 99% of the sequences passed MG-RAST internal QC; 23.2% and 23.4% of these sequences contained predicted proteins with known and unknown functions respectively.
Bacterial taxa overwhelmingly dominated the Rotmoosferner cryoconite metagenome, with over 83% of all contigs aligned at an e value of 1 × 10−5 or less to Bacteria within the M5NR database. Eukaryotes accounted for only 0.6%. Interestingly, while reports of the presence of Archaea in cryoconite are restricted to Antarctic glaciers (Cameron et al 2012b) and one cryoconite hole in the Rocky Mountains (Hamilton et al 2013), reads aligned to Archaea comprised a very small minority (0.1%) of the metagenome, and were predominantly Methanomicrobiales affiliated (2198 contigs of 2357 archaeal-affiliated contigs).
Considering that glacial environments often show high virus–bacterium ratios, the paucity of viral-associated contigs is surprising (Anesio and Bellas 2011). Considering the limited information on the genetic diversity of viruses (Anesio and Bellas 2011) from glaciers, their representation in databases may be limited, thus contributing to the residue of reads which could not be assigned to established taxa.
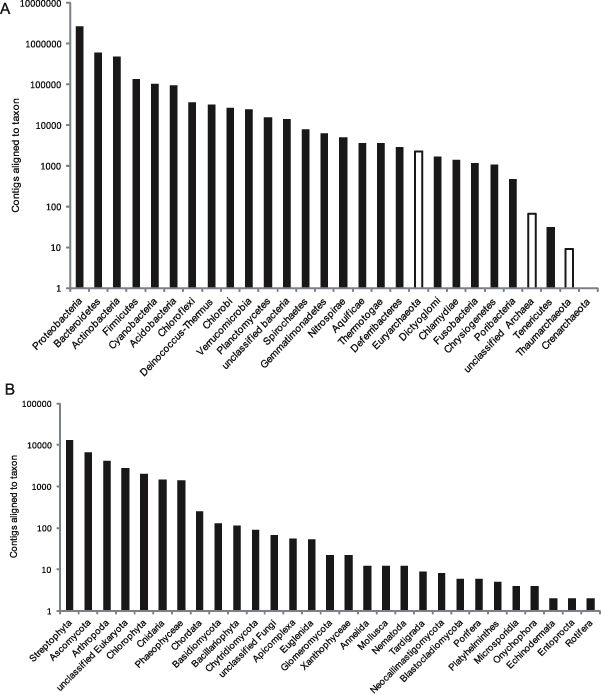
The bacterial affiliated contigs were distributed across 23 phyla (figure 2). Less than 1% of bacterial contigs were unassigned at the level of phylum, and the reads assigned to phyla ranged in abundance between 32 reads for Tenericutes and 2.6 × 106 reads affiliated to Proteobacteria. Correspondingly, the top five bacterial phyla were Proteobacteria (63.2% of bacterial reads), Bacteroidetes (14.0%), Actinobacteria (11.3%), Firmicutes (3.3%) and Cyanobacteria (2.5% of bacterial contigs). Within the five-most dominant bacterial phyla, several classes were present (figure 3). Of particular note is the abundance of Betaproteobacteria, which accounted for half (52%) of all Proteobacteria, with Alphaproteobacteria (24%) and other classes accounting for the remainder. The dominance of cryoconite bacterial communities by Proteobacteria has been reported in other locations (Cameron et al 2012b, Edwards et al 2011). In the Alps, the prominence of Betaproteobacteria within glacial ice (Simon et al 2009), and supraglacial debris on Rotmoosferner itself (Philippot et al 2011) have been described. It is therefore likely that Proteobacteria, and in particular Betaproteobacteria play important roles within the Rotmoosferner cryoconite communities. This is consistent with our understanding of Betaproteobacteria as early colonizing, r-selected taxa (as related by Fierer et al 2007, 2010, Pianka 1970, Zumsteg et al 2012). Moreover, Betaproteobacteria are prominent members of snowpack communities (Hell et al 2013, Larose et al 2010), and in alpine regions, Betaproteobacteria may play important roles in foreland mineral weathering (Frey et al 2010) and debris covered glaciers (Franzetti et al 2013). This raises the potential for cryoconite ecosystems to act as foci of biogeochemically reactive Betaproteobacteria which consequently inoculate glacier forelands.
Figure 2. Assignment of contigs at phylum level (e ≤ 1 × 10−5) for (A) Bacteria (black bars) and Archaea (white bars) and (B) Eukaryotes on the basis of MG-RAST analyses. Note the different scales for (A) and (B).
Download figure:
Standard image High-resolution imageFigure 3. Class level assignment of contigs (e ≤ 1 × 10−5) for the five-most dominant bacterial phyla in the cryoconite metagenome.
Download figure:
Standard image High-resolution imageInterestingly, four classes (plus unclassified) of Bacteroidetes were present in roughly equal proportion, implying phylum level ecological coherence (Philippot et al 2010) in cryoconite. This phylum is abundant in a diverse array of habitats, ranging from gastrointestinal tracts to the cryosphere, but typically appears well-adapted to the exploitation of recalcitrant polysaccharides (e.g. plant fibre; Thomas et al 2011). Their prevalence within this metagenome raises the possibility of involvement in the degradation of plant-derived organic matter. This is consistent with the prominence of allochthonous organic matter (Stibal et al 2008) and in particular the abundance of lower-order plant biomarkers (e.g. mosses and lichens) in Rocky Mountain cryoconite (Xu et al 2009).
Indeed, for habitats typically associated with significant rates of primary production (Anesio et al 2009), the limited abundance of Cyanobacteria in the Rotmoosferner cryoconite metagenome was surprising. Equally, eukaryotic photosynthetic taxa were rare (figure 2). Although the eukaryotic metagenome of Rotmoosferner cryoconite was dominated by Streptophyta, it can be assumed that these bryophytes represent legacy DNA within the cryoconite community. Consequently, the metagenome sequence may provide insight into sources of allochthonous organic matter as well as community composition. The presence of mosses derived from ice-marginal areas on the glacier surface (Joklamys Porter et al 2008) is well known and, aptly, these are apparent on Rotmoosferner, albeit in a less extravagant fashion, and in association with arthropods (figure 1(D)). The second-most dominant category of eukaryotic-affiliated contigs were the Ascomycota, which have been described on High Arctic glacier cryoconite (Edwards et al 2012). Fungi are obligate osmotrophic heterotrophs, although other, phototrophic eukarya (such as Chlorophyta) are present at lesser abundance. The scarcity of these eukaryotic phototrophs relative to eukaryotic heterotrophs and presumed legacy DNA from Streptophyta appears compatible with the net heterotrophic state revealed by process measurements.
In summary, the phylogenetic composition of the Rotmoosferner cryoconite metagenome lends support to the notion that the cryoconite ecosystem of Rotmoosferner is typified by aggregates of predominantly heterotrophic bacteria, minerals and allochthonous organic matter and contemporaneous measurements of net ecosystem productivity for Rotmoosferner cryoconites generally indicated net heterotrophy. It is therefore likely that (albeit, later in the season, on this north-facing glacier) the cryoconite ecosystem is typified by net heterotrophy and subsists on allochthonous organic matter, rather than net autotrophy, which is supported by the limited abundance of cyanobacterial and eukaryotic phototroph sequences.
To investigate the functionality of the Rotmoosferner cryoconite metagenome further, genes assigned to the Subsystems functional category hierarchy at an e value of 1 × 10−2 or less were analysed (figure 4(A)). Although the caveat that functional gene presence in glacial environments does not necessarily equate to functionality (Brankatschk et al 2010) applies to PCR detection of functional genes and metagenomics alike, many metabolic pathways were revealed (KEGG reconstruction: supplementary figure 1 available at stacks.iop.org/ERL/8/035003/mmedia), with complete glycolytic, TCA cycle, fatty and amino acid and nucleotide metabolism detected.
Figure 4. Distribution of Subsystems functional categories. A top level categories; bar represents the category mean relative abundance for the 32 metagenomes described in text with error bar ±1 SEM framed against cryoconite (diamonds). (B) Log scale abundance of contigs present in subsystems associated with macronutrient cycles.
Download figure:
Standard image High-resolution imageFirst, comparison to a subset of publically available metagenomes from comparable environments was sought. The relative abundance of contigs assigned to functional categories of the cryoconite metagenome was framed against thirty-two other metagenomes. These metagenomes vary by sequencing effort and platform, however broadly plausible patterns in relative abundance were revealed.
Direct comparison (figure 4(A)) between the relative abundance of functional categories present in metagenomes from other environments and the cryoconite metagenome sequence was broadly congruent in rank order, with clustering based subsystems, carbohydrates, amino acids and derivatives among the dominant category. Interestingly, genes assigned to photosynthesis and dormancy or sporulation were the rarest categories.
Some inconsistencies in rank order of categories between cryoconite and the comparison set were apparent. These include a relative functional enrichment of genes in the regulation and cell signalling, fatty acids, lipids and isoprenoids, stress response, aromatic compound metabolism, sulfur metabolism, motility and chemotaxis categories in the cryoconite metagenome.
Supplementary table 2 (available at stacks.iop.org/ERL/8/035003/mmedia) lists the assignments in enriched categories. Membrane transport demonstrated an abundance of contigs matching ABC transporters and a complement of protein secretion systems, while the fatty acids, lipids and isoprenoids category was predominantly represented by fatty acid degradation regulons and genes associated with the metabolism of long chain fatty acids (cf plant biomarkers detected by Xu et al (2009) in cryoconite) over biosynthetic pathways. Within the category of sulfur metabolism, the assimilation of both inorganic and organic sulfur was represented, mainly by ferredoxin and glutathione uptake and metabolism. Glutathione is involved in acclimation to redox stresses in Cyanobacteria (e.g. Cameron and Pakrasi 2010) and is a common thiol among Cyanobacteria and Proteobacteria (Masip et al 2006). Indeed, the category of stress response itself was dominated by subcategories associated with fluctuating temperature, oxidative and osmotic stress which may illustrate some of the stresses associated with life in an environment with frequent freeze-thaw cycles. Other sources of stress were apparent with enrichment in aromatic compound metabolism (figure 4(B)). Although the levels of xenobiotic contamination on Rotmoosferner is unclear, cryoconite assemblages on other alpine glaciers have to contend with anthropogenic contaminants (Margesin et al 2002). Indeed, the category of virulence, disease and defence was dominated by genes associated with heavy metal resistance (29 448 contigs of 59 285 in the category), including elements with anthropogenic radionuclides present in other alpine glaciers (Tieber et al 2009) and antimicrobial resistance genes (22 738 contigs), which have recently been demonstrated on glaciers worldwide (Segawa et al 2013).
Cryoconite has been considered a supraglacial locus of microbial activity and biogeochemical cycling on the basis of process measurements, predominantly on Arctic glaciers (e.g. Anesio et al 2009, 2010, Edwards et al 2011, Hodson et al 2007, Stibal et al 2009, Telling et al 2011, 2012). Functional gene PCR reveals the genetic potential for cryoconite C and N cycling (Cameron et al 2012a, Telling et al 2012), however these observations were limited to polar glaciers.
Genes associated with N, S, Fe and P cycling were apparent in the metagenome. Overall, a trend towards assimilative metabolism was noted in each of the four categories associated with these elemental cycles (figure 4(B)). Although N fixation and denitrification associated genes were present, contigs aligned with ammonia assimilation and ammonification were more abundant, indicative of a preference for ammonia mineralization and uptake and hence a cryoconite nitrogen economy centred upon ammonia recycling. Sulfur metabolism is described earlier, while phylogenetically diverse categories of Fe(II) and Fe(III) acquisition are present. The cycling of phosphorus was indicated, with an abundance of phosphate metabolism, uptake and transport (including the Pho regulon) genes aligning to the cryoconite metagenome. This appears consistent with a demand for phosphate, a limiting nutrient in Svalbard cryoconite at least (Stibal et al 2009). In summary, the cryoconite metagenome of Rotmoosferner reveals the importance efficient acquisition of allochthonous carbon and available nutrients under stressful conditions which may be expected on the surface of an alpine glacier.
Second, comparison of the overall functional composition of the Rotmoosferner cryoconite metagenome with other metagenomes was sought, using the set of 32 metagenomes from comparable habitats (supplementary table 1 available at stacks.iop.org/ERL/8/035003/mmedia). This was achieved by multivariate analyses of functional category relative abundances. Firstly, principal component analysis was performed on the relative abundance of reads present in each functional category, explaining 91.4% of variation in PC1-2 (figure 5(A)), and secondly hierarchical cluster analysis of Bray–Curtis similarities derived from fourth-root transforms of relative abundances was conducted (figure 5(B)).
Figure 5. Multivariate analyses of top level Subsystems functional category. (A) Principal components analysis of category relative abundances and (B) hierarchical cluster analysis of Bray–Curtis distances of category relative abundances (with fourth-root transform).
Download figure:
Standard image High-resolution imageBoth analyses revealed ecologically plausible clustering patterns; for example the ordination and clustering of soil habitats as desert and non-desert soils, with cold (polar) deserts indistinguishable from hot deserts clearly recapitulates differences revealed in the original analyses of these datasets (Fierer et al 2012). The Rotmoosferner cryoconite metagenome fell within the cluster comprised of aquatic habitats, and was most similar to the alpine ice metagenome sequences and microbial mat type habitats, including ice-shelf microbial mats, sludge from phosphate removal biofilms and a stromatolite metagenome. Congruent clustering of habitats typified by sedimentary biofilms such as the above, in particular those from cold environments, implies similar functions to exploit niches within these habitats as well as commonality in the challenges faced by organisms. Thus, it appears that the cryoconite metagenome was shaped by selective forces comparable to those of other cold-dwelling sediment-associated biofilms.
A key strategy for survival in such cold-dwelling sediment-associated biofilms appears to be the promotion of highly efficient nutrient scavenging and recycling, as exemplified by the Proteobacteria dominated ice-shelf microbial mat metagenomes (Varin et al 2010, 2012), consistent with our analysis of the cryoconite metagenome. Both habitat types are likely to require efficient (re)cycling of allochthonous C and nutrients under stressful physical, chemical and nutritional conditions.
The similarity between glacial ice and cryoconite metagenomes is striking. Simon et al (2009) described a metagenome from near-surface (deeper than 30 cm) glacial ice during the ablation season. Since surface-decontaminated glacial ice was directly filtered onto 0.2 μm membranes upon melting (Simon et al 2009), it is possible that small aggregates of aeolian debris and microbes (cf Irvine-Fynn et al 2012) as well as purely englacial microbial communities were present. Therefore, a bilateral translocation of microbe–mineral aggregates dispersed in glacial ice and coalesced in cryoconite holes may occur.
4. Conclusions
We report one of the larger environmental metagenome assemblies to date, and, to our knowledge, the first metagenome from cryoconite. As our analyses are limited to a single location and time, their interpretation as a metagenomic 'snapshot' of a cryoconite ecosystem is advocated. This invites the subsequent application of comparative approaches to resolve how the cryoconite metagenome changes in response to spatial, temporal and environmental variation. Robust experimental design will be essential to gain further insights (Knight et al 2012), as will linking metagenomics and other meta-omics approaches (Turnbaugh and Gordon 2008). We are currently exploiting the rapid advances in the capability and accessibility of post-Sanger sequencing platforms to do so. Inevitably, since the mapping of metabolic diversity on a global scale is envisaged (the Earth Microbiome Project; Gilbert et al 2010), as the glacial biome represents a major freshwater habitat, the application of metagenomics to glacial systems will undoubtedly grow.
Nevertheless, from our metagenomic 'snapshot', a clearer picture of alpine cryoconite ecosystem structure and functionality comes into focus. The cryoconite ecosystem of Rotmoosferner appears dominated by Proteobacteria, particularly Betaproteobacteria and that heterotrophs such as Bacteroidetes are prevalent, presumably consuming allochthonous organic matter such as the bryophyte detritus detected in the metagenome (or apparent in biomarker studies Xu et al (2009)). The genetic potential for contributions to several nutrient cycles is revealed. Overall, it is likely that cryoconite communities on alpine glaciers exist as efficient scavengers and recyclers of allochthonous carbon and nutrients similar to microbe–mineral aggregates from other icy environments (Varin et al 2010, 2012).
In conclusion, although we effectively advocate the transition from a metagenomic 'snapshot' to a metagenomic 'album' of cryoconite ecosystems, it is clear that the cryoconite metagenome presented herein reveals the genetic foundations of an ice-cold hot-spot of microbial diversity and activity within the glacial biome.
Acknowledgments
AE acknowledges grants from the Society for General Microbiology (SGM PFRV10/4) which was instrumental to this study as well as NERC grant NE/K000942/1 which supported further collaboration and data analysis. BS and AJH acknowledge AFO Forschungsförderung GLAC.LIFE to BS and all fieldworkers are grateful for the assistance of alpine Forschungstelle Obergurgl. All authors thank the IBERS Translational Genomics Facility and the anonymous reviewers which have improved this version of the manuscript.