Abstract
Super paramagnetic iron oxide nanoparticles (SPIONs) (∼12 nm) were synthesized as the magnetic core for an imprinted polymer (MIP) shell using 4-vinylpyridine as the functional monomer and trimethylolpropane trimethacrylate (TRIM) as the cross-linker, bringing the average size up to ∼45 nm. Five targets were imprinted—the Selective Androgen Receptor Modulators (SARMs) andarine, ligandrol and RAD-140; and the steroids estradiol and gestrinone. All MMIPs produced good selectivity when loaded with a non-target molecule, with all calculated selectivity factors above the 1.2 recommended threshold and also demonstrated good affinity/capacity. The rebinding of the target molecules from a complex matrix was also explored by using spiked river water samples. The SARMs-based MMIPs were able to rebind 99.56, 87.63 and 72.78% of their target molecules (andarine, ligandrol and RAD-140, respectively), while the steroidal-based MMIPs were able to rebind 64.54 and 55.53% of their target molecules (estradiol and gestrinone, respectively) at a nominal loading of 20 ≈μg in 50 mg of NPs. This work highlights the potential of these bi-functional materials for trace material clean-up of complex samples and/or subsequent analysis and opens up possibilities for further simple, rapid-to-synthesise materials for targeted clean-up.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 4.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
Introduction
A global health crisis is emerging as androgen misuse morphs from performance enhancement among athletes to image enhancement within the general population [1–3]. Ready access to androgenic substances through online marketplaces is exacerbating the problem. Negative side effects of androgen use include aggression, depression, liver toxicity, and heart issues [4–7]. Anabolic steroids are one class of molecules that have been widely used as performance enhancing drugs. Testosterone, the key androgen was first isolated in the 1930s and within the following year, hundreds of synthetic androgens were synthesized. Use of these compounds quickly became widespread by elite athletes to greatly improve muscle mass and athletic performance, leading to them being placed on the list of banned substances of the International Olympic Committee (IOC) in 1976 and 'out-of-competition' doping tests being introduced as many athletes used these steroids in their training period instead of during competition [8]. As anabolic steroid use spread from elite athletes to the general population, the vast majority (∼80%) of today's androgen users take these drugs for personal appearance, rather than athletic competition [9]. The large amount of non-athletic users will likely account for the majority of the future public health problems associated with steroid abuse [2].
Selective androgen receptor modulators (SARMs) are a unique class of androgen receptor ligands currently being misused as a performance and image enhancing drug, that are able to bind to androgenic receptors and display tissue-selective activation [10]. SARMs were designed as more selective analogs of androgenic drugs, offering the possibility of wider therapeutic applications than steroids [11]. In a Duchenne muscular preclinical model, SARMs molecules displayed an increase in muscle mass and protein synthesis levels, comparable to that of oxandrolone, but with minimal off-target side effects [12]. The anabolic effect of SARMs along with reduced androgenic side effects has led to great interest within the body building community, along with significant potential for abuse amongst competitive athletes [13]. SARMs molecules are readily available for purchase from a range of unverifiable sources without FDA approval. Due to the potential for abuse in both amateur and professional sport SARMs have been included in the prohibited substance list by the World Anti-Doping Agency (WADA) [14].
Wastewater-based epidemiology (WBE) is one method that is widely used to estimate illicit drug consumption within the general population, and also has the potential for monitoring performance and image enhancing drug use [15]. This principle is also used in environmental studies with river water to measure environmental spread and impact of drugs. While there have been limited studies that investigate the stability and degradation of SARMs in wastewater, there is an important need to be able to be able to extract and quantify these compounds from complex matrices, especially as they are becoming more widespread in use [16–18].
One approach is to employ antibodies or enzymes that offer strong affinity and high specificity [19–21]. These biological materials tend to be high cost, have long production times, specific conditions for use (pH, temperature, and ionic strength) and limited reusability [22, 23]. Development of synthetic recognition materials provides viable alternatives, especially where robustness is an important factor.
Molecularly imprinted polymers (MIPs) are a class of synthetic recognitions materials that have the potential to match the performance of their biological counterparts, while removing their downsides. MIPs have shown the ability to offer robustness in a variety of conditions, while offering a high degree of selectivity and specificity. MIPs are traditionally produced using a self-assembly method of functional monomers around a template (target analyte) forming a complex via non-covalent interactions [24]. The monomers are then polymerised, using a suitable cross-linker, entrapping the complex within the highly cross-linked polymer structure. After the removal of the template, the polymer is left with binding cavities, which are sterically and functionally complimentary to the template molecule [24].
The use of MIP nanoparticles (nanoMIPs) significantly improves MIP performance by generating more particle uniformity and better-defined binding sites, compared with the grinding of bulk MIPs to powder [25–28]. The sold-phase synthesis technique further enhances synthetic control and limits the number of binding sites per nanoparticle, enabling the nanoMIPs to offer excellent binding capacities and performances that are comparable to that of monoclonal antibodies [29–32]. Multi-step solid-phase synthesis has proven to be a popular technique, due to providing a pathway to high affinity nanoMIPs. Low yields and the need for functionalization of the solid support, immobilization of the target etc complicate the solid-phase approach. Many of these problems can be avoided using core–shell polymerization, where a solid core serves as a nucleation site for the template. Polymerisation yields a thin layer of MIP around the core [33] with good control of particle size. The core also adds unique characteristics such as density, magnetic or optical properties, further enhancing nanoMIP capabilities [33].
This study investigates the development of magnetic MIP (MMIP) nanoparticles, using a core–shell approach, for the SARMs targets andarine, ligandrol and RAD-140 (figure 1) and the application of these materials in rebinding these compounds from waste water. The core–shell approach was developed to incorporate magnetic properties within the imprinted material. The MIPs were characterised using Fourier-transform infrared spectroscopy (FTIR) and dynamic light scattering (DLS). Binding and selectivity of the MIPs was investigated using HPLC analysis.
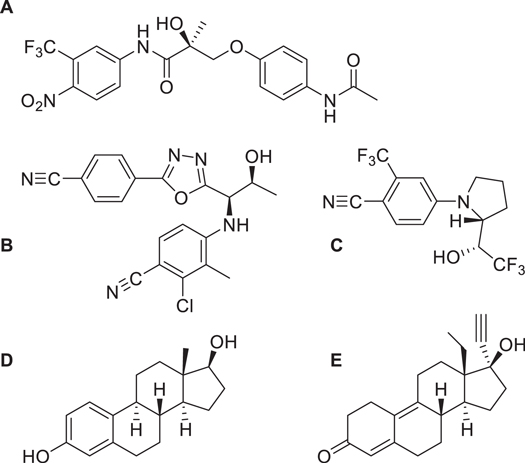
Figure 1. Structures of compounds involved in this study. (A) Andarine; (B) RAD-140; (C) Ligandrol; (D) Estradiol; and (E) Gestrinone.
Download figure:
Standard image High-resolution imageAlongside this SARMs study, we have also explored the same imprinting method for two steroidal targets, demonstrating that the method has flexibility for different classes of compounds. We have selected gestrinone (a medication used for treatment for endometriosis) and estradiol (a natural hormone used in menopausal hormone therapy), as these compounds are known to be present in river and waste water (figure 1) [16, 34, 35]. By imprinting two different classes of compounds we can explore the validity of the nanoparticle core–shell technique.
While steroids have been a common imprinting target for decades making them a model system [36–38], SARMs have only recently come to the fore with only a single study (a sensor platform developed by the Author's team) in the literature [25].
Method
Materials
All chemicals and solvents were analytical quality or high-performance liquid chromatography (HPLC) grade and were used as found without further purification.
4-vinylpyridine (4-VP), acetic acid, azobisisobutytonitrile (AIBN), chloroform, ethylene glycol, ethylene glycol dimethacrylate (EGDMA), iron (III) chloride, methacrylic acid (MMA), methanol, polyvinyl alcohol (PVA), sodium acetate (NaOAc), trimethylolpropane trimethacrylate (TRIM), were all purchased from Fisher Scientific UK (Loughborough, Leicester, UK). Andarine, Estradiol, Gestrinone, Ligandrol, and RAD-140 were purchased from Biosynth Carbosynth, Compton, Berkshire, UK.
Instrumentation
Microwave synthesis was carried out using a CEM Discover 2.0 with autosampler and the nanoparticles were sized using a Brookhaven Nanobrook Omni particle sizer, with double distilled water used as a dispersant. FTIR spectroscopy of the samples was undertaken using a Bruker FT-IR spectrometer (Alpha model) in ATR mode. MIP rebinding studies were performed using an Agilent HPLC 1100 series with diode array detector (DAD).
HPLC chromatographic conditions
An Agilent 1100 series HPLC was used to determine the adsorption performance of the MIPs. The sample was separated on a HypersilTM BDS C18 column (4.6 × 250 mm, particle size of 5 μm) The mobile phase consisted of 4:1 methanol:water with a flow rate of the mobile phase maintained at 1 ml min−1 and the column temperature was set at 35 °C. The injection volume was 5 μl. A DAD was used at wavelengths 248, 230, 345, 300 and 275 nm for andarine, estradiol, gestrinone, ligandrol, and RAD-140, respectively.
Synthesis of superparamagnetic iron oxide nanoparticles
Superparamagnetic iron oxide nanoparticles (SPIONs) were prepared using a one-pot solvothermal method, adapted from Sullivan et al [39]. With stirring, 0.5 g of FeCl3·6H2O and 1.5 g of NaOAc were dissolved in 15 ml of ethylene glycol in a 35 ml CEM Pyrex pressure vessel microwave reaction vial (MRV). The stirring continued for five minutes. The magnetic stirrer bar was then removed and the MRV was placed into a CEM Discover 2.0 (with autosampler) and the reaction was heated up to a temperature of 200 °C with a ramp time from 20 °C of two minutes. The reaction was held at 200 °C for 20 min under pressure (9 bar). The resulting composite products were washed five times with deionised water followed by two washes of methanol, and then collected with a magnet and finally dried in an oven at 60 °C for further use.
Magnetic molecularly imprinted polymer nanoparticle synthesis
To a solution of the selected template (0.05 mmol) in methanol (10 ml), 32 mg of the functional monomer 4-VP (0.30 mmol) was added and stirred for 30 min to form a template-monomer complex. Next, 25 mg of the SPIONs were added with 102 mg of the cross-linker TRIM (0.30 mmol). Finally, 12.5 mg of AIBN (0.08 mmol) was added. The solution was vortexed for 3 min, then purged with nitrogen for 5 min, before being sealed and place in an oven at 60 °C for 24 h. After polymerisation, the MMIPs were collected magnetically and washed with methanol/acetic acid (3:1 v/v) until no template was detected in the wash solutions by HPLC. Finally, the MMIPs were rinsed with methanol and left to dry for 24 h.
Rebinding studies
The rebinding ability of the magnetic molecularly imprinted polymers (MMIPs) was examined by the addition of 50 mg of the MMIP into an Eppendorf tube. Then 1 ml of a 20 μg ml−1 solution of the target molecule in methanol was added, with the mixture vortexed and left for one hour for rebinding to occur at room temperature. The MMIP was separated from the solution using a magnet, the supernatant was then filtered through Whatman (No. 1) filter paper. Remaining target left in the supernatant, after MMIP rebinding was measured using an Agilent 1100 series HPLC (C18 column set to a temperature of 35 °C and a mobile phase of methanol:water 4:1 at a flow rate of 1 ml min−1). A DAD detector was used with the appropriate wavelength set for the target; 248, 230, 345, 300 and 275 nm for Andarine, Estradiol, Gestrinone, Ligandrol and RAD-140, respectively. The amount of target analyte bound to the MMIP, after rebinding, was calculated by subtracting the final concentration of target in the supernatant from the known amount of target loaded.
Results and discussion
Synthesis of superparamagnetic iron oxide nanoparticles
A rapid microwave-assisted synthesis was successfully used to quickly and efficiently produce SPIONs. The method used was adapted from Sullivan et al with the microwave energy directly transferring to the reaction components that are susceptible to microwave absorption [39]. This improves efficiency by only heating the reaction mixture, reducing the need to heat reaction vessels, unlike conventional heating [40]. This allows the heating of the reagents to be much faster than conventional methods, thus minimising the time taken for the activation energy of the reaction to be reached. This fundamentally reduces reaction time, while also minimising unwanted side reactions.
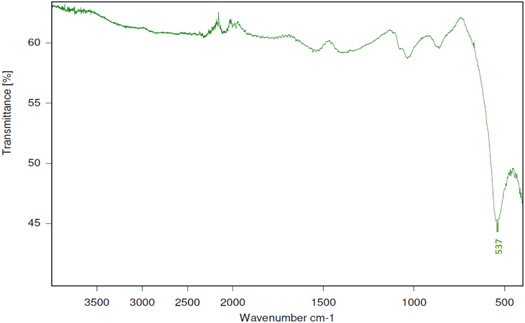
The FTIR-ATR spectra of the SPION is shown in figure 2 and shows an absorption band at 537 cm−1 which is assigned to the Fe-O stretching vibration and confirms that iron oxide particles have been produced through the one-pot microwave facilitated solvothermal method. With the exception of residual reaction solvent (ethylene glycol) peaks at approximately 800 and 1200 cm−1, the absence of any other major peaks, confirms that only iron oxide has been produced and is consistent with literature [39]. With limited by-products, this synthetic method demonstrates increased yield over other methods [40]. Furthermore, DLS (Figure S1) has shown that these particles have an average size of 11.8 (± 2.1) nm and is consistent with literature [39, 41]. A narrow size distribution is obtained, which is desirable for providing well defined coatings in later steps. It should be noted that SPIONs have wider applications in a variety of industries and this particular synthetic technique is rapid, controllable and exceptionally cost effective.
Figure 2. FTIR spectra of microwave-assisted SPION. The key peak at 537 cm−1 is highlighted. Obtained using a Bruker Alpha FT-IR spectrometer.
Download figure:
Standard image High-resolution imageMagnetic molecularly imprinted polymer nanoparticles synthesis
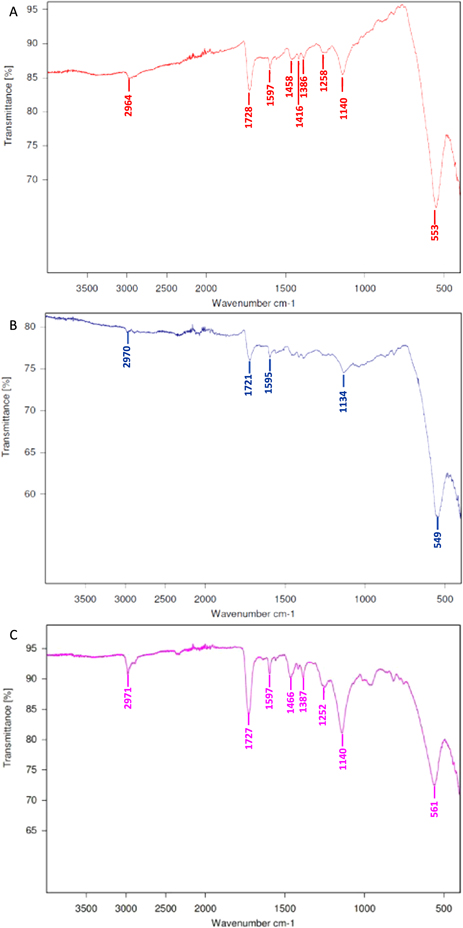
Using 4-VP as the functional monomer and TRIM as the cross-linker, a molecularly imprinted polymer layer, templated for the SARMs molecules andarine, ligandrol and RAD-140, was produced around the SPION. FTIR-ATR spectra for the magnetic MIPs are shown in figure 3, with figures 3(A)–(C) for andarine, ligandrol and RAD-140 MIPs, respectively and a summary of the absorption bands and their functional group assignments shown in table 1. The addition of absorption bands above 1000 cm−1, clearly show that a polymer layer has formed, when compared with figure 2. Furthermore, the absence of a C=C alkene peak (at approximately 1620–1610 cm−1) in all spectra confirms that the monomer (4-VP) and cross-linker (TRIM) have polymerised and the characteristic peaks shown are consistent with that of a polymer layer being formed around the SPION. It should be noted that the template/target molecule stretching bands (shown in figure S3), especially the strong/distinctive bands which would be expected to be visible, are absent from the spectra. This is consistent with literature and is possibly due to the template/target peaks being masked by bands form the polymer, especially with such a low amount of target/template compared with polymer [42].
Figure 3. FTIR spectra of the magnetic MIPs for the targets: Andarine (A), Ligandrol (B) and RAD-140 (C), obtained using a Bruker Alpha FT-IR spectrometer.
Download figure:
Standard image High-resolution imageTable 1. A summary of the FTIR absorption bands for the different MMIPs.
| FTIR bands (cm−1) | |||||
|---|---|---|---|---|---|
| MMIP | Fe–O | C–O (TRIM) | C=C cyclic (4-VP) | C=O (TRIM) | C–H |
| Andarine | 553 | 1140 | 1597 | 1728 | 2964 |
| Ligandrol | 549 | 1134 | 1595 | 1721 | 2970 |
| RAD-140 | 561 | 1140 | 1597 | 1727 | 2971 |
| Estradiol | 554 | 1139 | 1597 | 1724 | 2951 |
| Gestrinone | 540 | 1147 | 1543 | 1716 | 2981 |
The addition of the MIP layer resulted in an increase in nanoparticle size, with DLS now showing the nanoparticles to be 38.6 (±1.8) nm, 47.5 (±1.7) nm and 35.3 (±1.6) nm for the Andarine, Ligandrol and RAD-140 MIPs, respectively (Figure S4). This increase in size is to be expected as a polymer layer has now formed around the nanoparticle [33]. The slight differences between MMIP particle size are most likely due to the different template sizes, which act as an anchoring point for the functional monomers, forming a template-monomer complex via a self-assembly approach, within the pre-polymerisation solution. The functional monomers are then crosslinked together, forming an uneven polymeric layer around the magnetic core, producing MIP nanoparticles with a range of size distributions [19, 20, 25, 26]. It is generally considered that having much larger sized MIP particles, those in the μm size compared with nm, will decrease the binding ability of MIPs, due to smaller (nm sized) particles having a much bigger relative surface area, thus leading to more usable material [27, 31, 32]. The difference in size for the particles produced within this study is not considered significant enough to affect the rebinding of the target molecule, as demonstrated by previous work [20, 25]. After removal of the template through a series of methanol/acetic acid (3:1 v/v) washes, the particles were ready for rebinding studies.
Additional MMIPs were produced, in ordered to further explore their suitability as a recognition material for the collection of molecules of interest. This time the MMIPs were produced in the same manner, but for the steroidal targets estradiol and gestrinone. FTIR spectra for the magnetic MIPs are shown in figure Figures S2(A) and (B) (estradiol and gestrinone, respectively) and a summary of the absorption bands and their functional group assignments also included in table 1. Again, the addition of absorption bands above 1000 cm−1 demonstrations that a polymer layer has been formed around the SPIONs. Polymerisation of the monomer (4-VP) and cross-linker (TRIM) is also confirmed again with the absence of peaks at approximately 1620–1610 (C=C alkene). The template/target molecule stretching bands (shown in figure S3), are also absent from the spectra and is possibly due to the template/target peaks being masked by bands from the polymer, especially with such a low amount of target/template compared with polymer [42]. Again, addition of the MIP layer increased the nanoparticle size as shown by DLS, the nanoparticles were 42.2 (±2.0) nm and 51.0 (±1.9) nm for the estradiol and gestrinone templates (Figure S5). After full removal of the template through a series of methanol/acetic acid (3:1 v/v) washes, the particles were ready for rebinding studies.
Magnetic molecularly imprinted polymer nanoparticles rebinding studies
The rebinding performance was measured by utilising a subtraction technique, whereby a known concentration of the target was mixed with the MIP and allowed to associate. Here we used methanol as the solvent due to the target compounds having mixed solubility profiles. This is rare as methanol is often used to wash the polymer as its polar nature commonly breaks the ionic interactions between template and monomer (the strength and nature of interactions are naturally template-monomer dependent). However, in this work we observed that methanol on its own was not strong enough to do this and acidified methanol was required [33]. The supernatant was then analysed using an Agilent 1100 series HPLC fitted with a DAD detector, and the amount of target bound was calculated. An initial calibration was plotted by injecting known concentrations (0.5–50 μg ml−1) of the target molecules (dissolved in methanol and passed through filter paper), then plotting signal response (peak area) over concentration (Figures S6(A)–(E)). The percentage rebinding of the target to the MMIPs are shown in Figures 4(A)–(C), for the Andarine, Ligandrol and RAD-140 MIPs, respectively and a summary shown in table 2.
Figure 4. Percentage of SARMs rebinding to individual imprinted MMIPs. (A) Andarine MMIP. (B) Ligandrol. (C) RAD-140. N = 3. Sample from 1 ml of a 20 μg ml−1 solution of the target molecule.
Download figure:
Standard image High-resolution imageTable 2. Percentage rebinding and selectivity factors for different SARM imprinted MMIPs.
| Percentage target bound (%) | Selectivity factors (SF) | |||||
|---|---|---|---|---|---|---|
| Andarine | Ligandrol | RAD-140 | Andarine | Ligandrol | RAD-140 | |
| Andarine | 99.20 (±0.01) | 40.06 (±0.02) | 49.99 (±0.01) | — | 2.48 | 1.98 |
| Ligandrol | 62.68 (±0.48) | 87.14 (±0.10) | 61.06 (±0.13) | 1.39 | — | 1.43 |
| RAD-140 | 44.06 (±0.15) | 43.95 (±1.17) | 75.19 (±1.43) | 1.71 | 1.71 | — |
Traditionally, the use of a non-imprinted polymer (NIP) was seen as a way to measure the strength of the interaction between the MIP and the target molecule [24]. However, multiple studies have shown that the NIP has significantly different morphology and behaviour to a MIP [43]. The presence of the template can affect the rate of reaction, and the porosity. It is generally accepted that using a selectivity factor SF is considered more favourable and provides a better overall measure of the binding ability of the MIP [44]. This is calculated using equation (1), where target analyte binding is compared to non-target analyte. The selectivity of the MMIPs were explored further by studying their binding with non-target SARMs molecules, chosen due to similarity in size and structure.
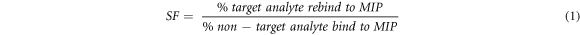
SF values greater than 1.2 considered acceptable [39], and a demonstration of imprinting [42]. The SF values for the MMIPs are shown in table 2. As shown by table 2 the Andarine MMIP has SF values of 2.48 and 1.98, when loaded with the non-targets Ligandrol and RAD-140 respectively. These are good SF values, and higher than the 1.2 threshold. This shows that the Andarine MMIP is specifically selective for the target molecule Andarine and not for the other non-targets. Given the difference in structures (figure 1) this is not surprising as the 'pocket' will have a different shape profile. The Ligandrol MMIP has SF values of 1.39 and 1.43, when loaded with the non-targets Andarine and RAD-140, respectively. Whilst these SF values are not as good as those for the Andarine MMIP, they are still above the recommended 1.2 threshold, thus showing the Ligandrol MMIP still has a reasonable degree of selectivity. Finally, the RAD-140 MMIP has better SF values (than the Ligandrol MMIP), with values of 1.71 for the non-target loading of Andarine and Ligandrol again showing that this MMIP is selective for its the target.
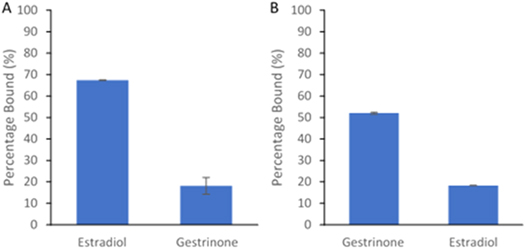
This core–shell MIP approach was investigated again, but with the steroidal-based target molecules, estradiol and gestrinone. As figure 5(A) and table 3 shows, the estradiol MMIP rebinds 67% of the target, when loaded with estradiol and 18% of the non-target, when loaded with gestrinone. Figure 5(B) (and table 2) shows the gestrinone MMIP is able to rebind 52% of the target when loaded with gestrinone and ∼18% of the non-target estradiol.
Figure 5. Percentage of rebinding to the different steroidal imprinted MMIPs. (A) Estradiol imprinted MMIP. (B) Gestrinone imprinted MMIP. N = 3. Sample from 1 ml of a 20 μg ml−1 solution of the target molecule.
Download figure:
Standard image High-resolution imageTable 3. Percentage rebinding and selectivity factors for different steroidal imprinted MMIPs.
| Percentage target bound (%) | Selectivity factors (SF) | |||
|---|---|---|---|---|
| Estradiol | Gestrinone | Estradiol | Gestrinone | |
| Estradiol | 67.41 (±0.14) | 18.19 (±3.89) | — | 3.71 |
| Gestrinone | 18.32 (±0.09) | 52.06 (±0.53) | 2.84 | — |
Whilst, the overall percentage rebinding for these steroidal-based MMIPs are lower (table 3) than their SARM MMIPs counterparts (table 2), their selectivity factor (SF) values, presented in table 3, are much higher. The estradiol MMIP has a SF value of 3.71, when loaded with gestrinone, and the gestrinone MMIP has a SF value of 2.84, when loaded with estradiol. This shows that both MMIPs have excellent selectivity towards their chosen target and not towards other molecules, even those of from the same class.
The MMIPs produced in this study, follow the adaptation of a generalised synthesis method, whereby a non-target specific functional monomer (4-VP) is used. This results in the range of the amount (percentage) of target able to rebind to the MMIP, presented in tables 1 and 2, and is consistent with the work of El-Sharif et al, who also showed differences with the percentage of target rebinding with MIPs formed using the same functional monomers [45]. As such the comparison of the MIP against a control (NIP or Selectivity) is a much better measure of the selectivity of the MIP [42].
Extraction from water samples
As highlighted in the introduction, the ability to selectively rebind these families of compounds from water is important for understanding community drug use through WBE and other environmental tracing. Therefore, we repeated the extraction using river water samples This was collected from the River Soar on the 3rd March 2022 at co-ordinates 52°37'51.2''N, 1°08'32.7''W. The collected water was filtered through a 0.22 um filter in order to remove any sediment and/or any organic matter (bacteria etc), and then spiked with either the SARMs or steroidal compounds at 20 μg ml−1. 1 ml of the spiked samples were mixed with 50 mg of the corresponding MMIP, and the amount of target bound to the MMIPs was calculated after the polymer was collected using a magnet and measured through an extraction method.
The percentage of the target analyte bound to the MMIP is summarised in table 4. The MMIPs are able to rebind and collect a very high percentage of their imprinted target from river water samples, which is consistent with the amount of target rebound in the initial model studies. This shows that the complex media of the river water samples does not have any interfering effect on the media, allowing the MMIPs to bind analytes within complex media.
Table 4. Percentage rebinding of SARMs/steroidal target to their corresponding MMIPs from a river water sample (1 ml of 20 μg ml−1 solution with 50 mg of polymer. N = 3.
| Magnetic MIP | Percentage target bound (%) |
|---|---|
| Andarine | 99.56 (±0.25) |
| Ligandrol | 87.63 (±20.9) |
| RAD-140 | 72.78 (±0.68) |
| Estradiol | 64.54 (±0.90) |
| Gestrinone | 55.53 (±1.49) |
Conclusions
Here we have demonstrated the selective recognition of series of magnetic molecularly imprinted polymer nanoparticles for the selective recognition of three SARMs as well as two steroidal molecules.
Using a microwave solvothermal method a superparamagnetic iron oxide nanoparticle was produced, which was then used as a magnetic core for the MMIP. A molecularly imprinted polymer shell was then synthesized around the SPION, in a core–shell approach using 4-VP as the functional monomer and TRIM as the cross-linking agent, to produce a MMIP nanoparticle. The magnetic properties of the materials allow them to be easily separated from the solution under study. The MMIPs produced were shown to exhibit good capacity and selectivity for their target molecules when tested with other compounds in their class. Selectivity factors for all polymers were over the recommended 1.2 threshold ratio. They also were able rebind the compounds from the complex media of river water highlighting potential applications in analytical methodologies.
This simple study is a proof-of-concept work that demonstrates the potential of these easy-to produce bi-functional materials. The focus of our follow-on studies is to expand and improve the performance of the target imprinted polymer shells for these useful MMIP materials. In this work, we have used a single monomer-cross linker system for all targets, and we are looking to apply molecular modelling approaches to select polymer composition to improve the selectivity and affinity. We are also exploring the polymerisation reaction conditions to study the thickness of the polymer shell and size distribution towards controllable size. Further studies using different matrices, targets and analytical instrumentation to improve the sensitivity are underway. We envisage that these bi-functional nanomaterials that offer both chemical selectivity and magnetic properties will play an interesting part in future analytical methodology where solid phase extraction is not suitable. Likewise, the size of these materials (in the nanometre scale) suggests that they could be used for labelling.
Acknowledgments
BD wishes to thank Professor Anwar Baydoun and the Faculty of Health & Life Sciences graduate program for financial support. AM, BF, CF, MG, RP wish to thank the School of Pharmacy undergraduate project programme for financial support.
Data availability statement
All data that support the findings of this study are included within the article (and any supplementary files).
Conflicts of interest
There are no conflicts to declare.
Supplementary data (0.5 MB PDF)