Abstract
The discovery of novel materials and functional molecules can help to solve some of society's most urgent challenges, ranging from efficient energy harvesting and storage to uncovering novel pharmaceutical drug candidates. Traditionally matter engineering–generally denoted as inverse design–was based massively on human intuition and high-throughput virtual screening. The last few years have seen the emergence of significant interest in computer-inspired designs based on evolutionary or deep learning methods. The major challenge here is that the standard strings molecular representation SMILES shows substantial weaknesses in that task because large fractions of strings do not correspond to valid molecules. Here, we solve this problem at a fundamental level and introduce SELFIES (SELF-referencIng Embedded Strings), a string-based representation of molecules which is 100% robust. Every SELFIES string corresponds to a valid molecule, and SELFIES can represent every molecule. SELFIES can be directly applied in arbitrary machine learning models without the adaptation of the models; each of the generated molecule candidates is valid. In our experiments, the model's internal memory stores two orders of magnitude more diverse molecules than a similar test with SMILES. Furthermore, as all molecules are valid, it allows for explanation and interpretation of the internal working of the generative models.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 4.0 license. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
The rise of computers enabled the creation of the field of computational chemistry and cheminformatics which deals with the development and application of methods to calculate, process, store and search molecular information on computing systems. Arising challenges of molecular representation and identification were addressed by SMILES (Simplified Molecular Input Line Entry System), which was invented by David Weiniger in 1988 [1]. SMILES is a simple string-based representation which is based on principles of molecular graph theory and allows molecular structure specification with straightforward rules. SMILES has since become a standard tool in computational chemistry and is still a de-facto standard for string-based representing molecular information in-silico.
Apart from predicting molecular properties with high accuracy, one of the main goals in computational chemistry is the design of novel, functional molecules. Exploring the entire chemical space–even for relatively small molecules–is intractable due to the combinatorial explosion of possible and stable chemical structures [2–4]. Substantial recent advances in artificial intelligence and machine learning (ML), in particular, the development and control of generative models, have found their way into chemical research. There, scientists are currently adapting those novel methods for efficiently proposing new molecules with superior properties [5–10]. For identifying new molecules, input and output representations are in many cases SMILES strings. This, however, introduces a substantial problem: A significant fraction of the resulting SMILES strings do not correspond to valid molecules. They are either syntactically invalid, i.e. do not even correspond to a molecular graph, or they violate basic chemical rules, such as the maximum number of valence bonds between atoms. Researchers have proposed many special-case solutions for overcoming these problems. For example, by adapting the machine learning models such that they deal with invalidity [11, 12]. While this solves the problems for specific models, it does not provide a universal solution for all current (and future) possible models. A different solution is to changing the definition of SMILES itself. This approach has been put forward in the work by O'Boyle and Dalke denoted DeepSMILES [13]. DeepSMILES could also be used as a direct input for arbitrary machine learning models and first raised the question of what the ideal string-based representation of molecules might be for generative tasks. DeepSMILES overcomes most synthactical issues to generate graphs, however, it does not deal with semantic constraints that are introduced by the specific domain. Thus, more than 30 years after Weininger's invention of SMILES, the applications of generative models for the de-novo design of molecules would benefit from a new way to describe molecules on the computer.
Here, we present SELFIES (SELF-referencIng Embedded Strings), a string-based representation of molecular graphs that is 100% robust. By that, we mean that each SELFIES corresponds to a valid molecule, even entirely random strings. Furthermore, every molecule can be described as a SELFIES. SELFIES are independent of the machine learning model and can be used as a direct input without any adaptations of the models.
We compare SELFIES with SMILES ML-based generative models such as in Variational Autoencoders (VAE) [14] and Generative Adversarial Networks (GANs) [15]. We find that the output is entirely valid and the models encode orders of magnitude more diverse molecules with SELFIES than with SMILES. Those results are not only significant for inverse-design of molecules, but also interpretability of the inner workings of neural networks in the chemical domain.
2. String-based representations of molecules
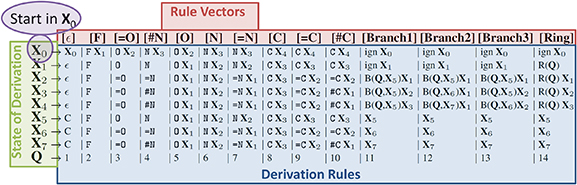
We are describing the string-based representations of SMILES and SELFIES using the small biomolecule 3,4-Methylenedioxymethamphetamine (MDMA). The SMILES string in figure 1(A) describes a sequence of connected atoms (green). Brackets identify branches and, and numbers identify ring-closures at the atoms that are connected. In SELFIES, figure 1(B), the information of branch length as well as ring size is stored together with the corresponding identifiers Branch and Ring. For that, the symbol after the Branch and Ring stands for a number that is interpreted as lengths. Thereby, the possibility of invalid syntactical string (such as a string with more opening than closing brackets), is prevented. Furthermore, each SELFIES symbols is generated using derivation rules, see table 2. Formally, the table corresponds to a formal grammar from theoretical computer science [18]. The derivation of a single symbol depends on the state of the derivation  . The purpose of these rules is to enforce the validity of the chemical valence-bonds.
. The purpose of these rules is to enforce the validity of the chemical valence-bonds.
Figure 1. Description of a molecular graph (of the small organic molecule, 3,4-Methylenedioxymethamphetamine) with two computer-friendly, string-based methods. (A) Derivation of the molecular graph using SMILES. The main string (green) is augmented with branches (defined by an opening and a closing bracket) and rings (defined by unique numbers after the atoms that are connected). Note that both branches and rings are non-local operations. (B) Derivation of the molecular graph using SELFIES. The main string is derived using a rule set such that the number of valence bonds per atom does not exceed physical limits. The symbol after a Branch is interpreted as the number of SELFIES symbols derived inside the branch. Its corresponding numerical value is explained in figure 2. The symbol after Ring interpreted as a number too, indicating that the current atom is connected to the (Q + 1)st previous atom. Thereby every information in the string (except the ring closure) is local and allows for efficient derivation rules.
Download figure:
Standard image High-resolution imageFigure 2. Derivation rules of SELFIES for small organic molecules. We denote SELFIES symbols as enclosed within brackets, and SMILES symbols without brackets. Every symbol of SELFIES is interpreted as a rule vector (top red line). A SELFIES symbol will be replaced by the string at the intersection of the rule vector and derivation state of the derivation (left, green). The string can contain an atom or another state of derivation. The derivation starts in the state  (violet), and continues in the state previously derived. The state of derivation takes care of syntactical and chemical constraints, such as the maximal number of valence bonds. The rules in state
(violet), and continues in the state previously derived. The state of derivation takes care of syntactical and chemical constraints, such as the maximal number of valence bonds. The rules in state  for n = 1-n = 4 are designed such the next atom can use up to n valence bonds.
for n = 1-n = 4 are designed such the next atom can use up to n valence bonds.  stands for function, creating a branch in the graph using the next Q symbols and starting in state
stands for function, creating a branch in the graph using the next Q symbols and starting in state  . R(Q) stands for a function that creates rings, from the current atom to the (Q + 1)-st previously derived atom. In both cases, the letter subsequent to R or B is interpreted as a number Q, which is defined in the last line of the table. The symbol
. R(Q) stands for a function that creates rings, from the current atom to the (Q + 1)-st previously derived atom. In both cases, the letter subsequent to R or B is interpreted as a number Q, which is defined in the last line of the table. The symbol  stands for an empty string, and ign means that the subsequent SELFIES symbol is ignored. This table covers all non-ionic molecules in the database QM9 [16, 17]. Ions, stereochemistry and larger molecules can also be represented by simply extending this table.
stands for an empty string, and ign means that the subsequent SELFIES symbol is ignored. This table covers all non-ionic molecules in the database QM9 [16, 17]. Ions, stereochemistry and larger molecules can also be represented by simply extending this table.
Download figure:
Standard image High-resolution imageAs a simple example, the SELFIES string [F][=C][=C][#N] is derived to SMILES in the following way. Here and everywhere else in the manuscript, we denote SELFIES symbols as enclosed within brackets and SMILES symbols without brackets. Starting in the state  , the first symbol (rule vector) [F] leads to F
, the first symbol (rule vector) [F] leads to F
 . The derivation of the second symbol subsequently continues in the state
. The derivation of the second symbol subsequently continues in the state  . The total derivation is given by
. The total derivation is given by
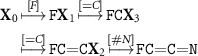
The final molecule FC=C=N, which satisfies all valence-bond rules, is 2-Fluoroethenimine. At this point, valence-bond constraints are satisfied for subsequent atoms and branches. The only remaining potential sources of violation of these constraints are the destination of rings. Therefore, we insert rings only if the number of valence-bond at the target has not yet reached the maximum. Thereby, using the rules in table 2, 100% validity can be guaranteed for small biomolecules. It is straight forward to extend the coverage for broader classes of molecules, as we describe below.
The derivation rules in table 2 are generated systematically and could be constructed fully automatically just from data, as we show in the Supplementary Information (SI), which is available online at https://stacks.iop.org/MLST/1/045024/mmedia. Furthermore, SELFIES are not restricted to molecular graphs but could be applied to other graph data types in the natural sciences that have additional domain-dependent constraints. We give an example, quantum optical experiments in physics with component dependent connectivity [19], in the SI.
Informal conversations with several researchers lead to the argument that SMILES are 'readable'. Readability is in the eye of the beholder, but needless to say, SELFIES are as readable as figure 1(B) attests to. After a little familiarity, functional groups and connectivity can be inferred by human interpretation for small molecular fragments.
3. Effects of random mutations
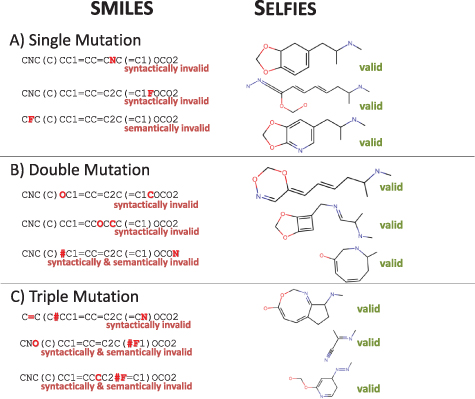
The simplest way to compare robustness between SMILES and SELFIES is by starting from a valid string, such as MDMA in figure 1, and introduce random mutations of the symbols of the string. In figure 3(A), we show three examples of one randomly introduced string mutation. We evaluate the resulting validity using RDKit [20]. All three SMILES strings are invalid. The first one is missing a second ring-identifier for 2, the second one is missing a closing bracket for a branch, and the last one violates valence-bond numbers of Fluorine. In contrast to that, all mutated SELFIES correspond to valid molecules. We can analyse one specific mutation, which changes the structure of SELFIES, specifically the middle SELFIES graph in figure 3(B). There, starting from the SELFIES string in figure 1(B), the first [Ring1] (purple) is replaced by a [#N]. As a consequence, the purple ring is not introduced, but a double-bond to a nitrogen atom is added to the main string, leading to the derivation state  . Besides, the symbol which indicates ring size ([#N], which stands for Q = 4) is now derived as a normal atom. As the current state is
. Besides, the symbol which indicates ring size ([#N], which stands for Q = 4) is now derived as a normal atom. As the current state is  , only a single bond (not a triple bond) to a nitrogen atom is introduced, and the derivation is concluded. This example shows the more intricate way how SELFIES does not allow for invalid molecules.
, only a single bond (not a triple bond) to a nitrogen atom is introduced, and the derivation is concluded. This example shows the more intricate way how SELFIES does not allow for invalid molecules.
Figure 3. Random Mutations of SMILES and SELFIES of the molecule in figure 1(A). (A) Single mutations have led to three invalid SMILES strings, while all SELFIES produce valid molecules. In (B) and (C) the initial molecule is two and three times mutated, respectively. In all cases, SMILES strings are invalid, while SELFIES produce valid molecules that deviate more and more from the initial molecule.
Download figure:
Standard image High-resolution imageIn figures 3(B) and (C), we introduce two and three mutations, respectively. Again, all SMILES are invalid, and all SELFIES are valid molecules. In general, the validity probability for SMILES with one mutation starting from MDMA is 26.6%, 9,0% and 3.7% for one, two and three mutations, respectively. DeepSMILES [13] improved these values to find that single, double and triple mutations for MDMA lead to 58,9%, 35,8% and 24,3% validity. Finally–SELFIES, are valid in 100% of the cases. Three examples for each case can be seen on the right panel of figure 3, details are in table 1.
Table 1. Results for bitflip (starting from the valid MDMA graph, using only involved tokens), random sequence, VAE and GAN.
| 1 bitflip | 10 bitflips | VAE | GAN | |||
|---|---|---|---|---|---|---|
| Validity | Validity | Validity | Reconstruction | Diversity | Diversity | |
| SMILES | 26.6% | 0.2% | 71.9% | 66.2% | 5.9% | 18.5% |
| DeepSMILES | 58.9% | 4.7% | 81.4% | 79.8% | 67.3% | − |
| SELFIES | 100% | 100% | 100% | 98.2% | 82.9% | 78.9% |
4. Results for deep generative models
Generative models are an ideal application of a 100% robust representation of molecules. One prominent example is a variational autoencoder (VAE) [14], which has recently been employed for the design of novel molecules [21]. In the domain of chemistry, the VAE is used to transform a discrete molecular graph into a continuous representation which can be optimized using gradient-based or Bayesian methods. As shown in figure 4, it consists of two neural networks, the encoder and decoder. The encoder takes a string representation of the molecule and encodes it into a continuous, internal representation (details about encoding and decoding can be seen in the SI). There, every molecule corresponds to a location in a high-dimensional space. The number of neurons defines the dimension in the latent space. The decoder takes a position in the latent space and transforms it into a discrete molecule (for instance again, a one-hot encoding of SMILES or SELFIES).
Figure 4. Variational Autoencoder (VAE) for Chemistry. The VAE is a deep neural network that takes a molecule as an input, encodes it to continuous latent space, and reconstructs it from there with a decoder. The latent space is a high-dimensional space where each point can be decoded into a discrete sequence. In contrast to a standard autoencoder, the latent neurons (green) encode Gaussian distributions (mean and variance), and the outputs are sampled from that distributions. We represent the molecular graphs using one-hot encodings of SMILES and SELFIES.
Download figure:
Standard image High-resolution imageThe goal of a VAE is learning to reconstruct molecules. After the training, one can scan through the latent space for optimizing chemical properties. Details can be found in table 1. Once an optimal point is identified, the decoder can map it to a molecular string. For any application of VAEs in chemistry, it is desirable that all points in the latent space correspond to valid molecules.
We experiment with a standard VAE, which we train to reconstruct molecules from the benchmark dataset QM9 [16, 17]. We employ both SMILES and SELFIES for that task. After the training, we analyze the validity of the latent space. We do this by sampling latent space points from randomly oriented planes in the high-dimensional space. Using SMILES, we find in figure 5(A) that only a small fraction of the space corresponds to valid molecules. A large fraction decodes to syntactically or semantically invalid strings that do not stand for molecules. In contrast to that, using SELFIES, we can see in figure 5(B) that the entire space corresponds to valid molecules. We want to stress that a 100% valid latent space is not only significant for inverse-design techniques in chemistry, but is essential for model interpretation [24–26], in particular for interpreting the internal representations [22, 23] in a scientific context [27]. The intuition is that it might be difficult for a human to conceptualize the meaning of regions that lead to unphysical molecular structures. Visualization techniques (which might help in understanding what the model has learned) that connect internal representation with physical properties cannot provide any insights in the invalid regions, thus lead to many scattered areas of valid molecules. It is an important open question whether the 100% valid latent space will indeed provide useful insights.
Figure 5. Validity of latent space. We analyze the latent space of a VAE, which was trained to reproduce small organic molecules from the QM9 database. The latent space has 241 dimensions (zi stand for a specific latent dimension). Upper row: We chose four randomly oriented planes in the high-dimensional space that go through the origin. Along the plane, we decode latent space points and calculate whether they correspond to valid or invalid molecules. The color code stands for the proportion of valid molecules (red = 0%, green = 100% valid). Lower row: We chose a random orientation of the plane, and displace it by a third random orientation by (−2, −1, +1, +2) standard deviations from the origin. In all experiments, we find that only a small fraction of the latent space for SMILES are valid, while for SELFIES the entire latent space is valid. This is not only important for generative tasks but is crucial for interpreting internals representations of the neural networks. Large and scattered invalid regions in the latent space might add challenges when interpreting the chemical properties of the output depending on internal regions; this issue is solved with SELFIES and can be exploited in future works [22, 23].
Download figure:
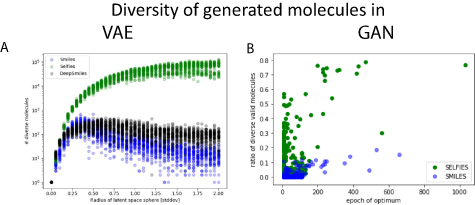
Standard image High-resolution imageBesides 100% validity, the molecule density in the latent space is of crucial importance too. The more valid, diverse molecules are encoded inside the latent space, the richer the chemical space that can be explored during optimization procedures. In figure 6(A), we compare the richness of the encoded molecules when a VAE is trained with SMILES and with SELFIES. For that, we sample random points in the latent space and stop after 20 samples did not produce any new molecule. We find that the latent space of the SELFIES VAE is more than two orders of magnitude denser than the one of SMILES.
Figure 6. Diversity of generative models trained with string-based representations, with the example of VAE and GAN. Beside robustness, diversity is one of the main objectives for generative models because it indicates the potential space in which optimization for chemical features occures. A) We investigate the density of valid diverse molecules by sampling the latent space of a VAE. We chose points with a distance of σ around the centre, stopping after 20 samples did not produce new instances. We find that the VAE trained with SELFIES contains 100 times more valid diverse molecules than if it is trained with SMILES or DeepSMILES. B) We train a GAN with 200 different hyperparameters to produce de-novo molecules for SELFIES and SMILES. Sampling 10.000 times, SELFIES produced 7889 different valid molecules, while for SMILES the most diverse valid number of molecules where 1855). Both cases show that SELFIES leads to significantly larger densities of diverse molecules compared to SMILES.
Download figure:
Standard image High-resolution imageOther prominent deep generative models are Generative Adversarial Networks (GANs) [15], which have been introduced in the design of molecules [28]. There, two networks–called generator and discriminator–are trained in tandem. The setting is such that discriminator receives either molecule from a dataset or outputs of the generator. The goal of the discriminator is to correctly identify the artificially generated structures, while the goal of the generator is to fool the discriminator. After the training, the generator has learned to reproduce the distribution of the dataset. We train the GAN, using 200 different hyperparameter settings both for SMILES and SELFIES. After the training, we sample each of the models 10.000 times and calculate the number of unique, valid molecules. For the best set of hyperparameters, we find that a GAN trained with SELFIES produces 78.9% diverse molecules while a GAN that produces SMILES strings only results in 18.6% diverse molecules, see figure 6(B).
5. Covering the chemical Universe
In this manuscript, we demonstrate and apply SELFIES for small biomolecules. However, the language can be extended to cover much richer classes of molecules. In the corresponding GitHub repository, we extend the language to allow for molecules with up to 8000 atoms per ring and branch, we add stereochemistry information, ions as well as unconstrained unspecified symbols. Thereby, we encoded and decoded all 72 million molecules from PubChem (the most complete collection of synthesized molecules) with less than 500 SMILES chars, demonstrating coverage of the space of chemical interest.
6. Standarization outlook
The SELFIES concept still requires work to become a standard. Upon publication of this article, the authors will call for a workshop to extend the format to the entire periodic table, allow for stereochemistry, polyvalency, aromaticity, isotopic substitution and other special cases so that all the features present in SMILES are available in SELFIES. Unicode will be employed to create readable symbols that exploit the flexibility of modern text systems without restricting oneself to ASCII characters. In that context, we will pursue to define direct canonicalization of SELFIES, such that there is a canonical SELFIES string for a unique molecule. Currently, SMILES can be made canonical indirectly, by translating them to SELFIES and convert the canonical SMILES back to SELFIES.
7. Conclusion
We presented SELFIES, a human-readable and 100% robust method to describe molecular graphs in a computer. These properties lead to superior behaviour in inverse design tasks for functional molecules, based on deep generative models or genetic algorithms. SELFIES can be used as a direct input into current and even future generative models, without the requirement to adapt the model. In generative tasks, it leads to a significantly higher diversity of molecules, which is the main objective in inverse design. In addition to the results presented here, in separate work, we use Genetic Algorithms and find that without any hard-coded rules, SELFIES outperform literature results in a commonly-used benchmark [29]. Apart from superior behaviour in inverse design, a 100% valid representation is also a sufficient condition for interpreting the internal structures of the machine learning models [27]. While we have focused on an representation that is ideal for computers, attention should also be drawn to SELFIES standardization to allow general readability [30], by exploiting the numerous remaining degrees of freedom of SELFIES.
Code and data availability
The full code is available at GitHub: https://github.com/aspuru-guzik-group/selfies. The dataset QM9, which has been used in this study, is available in reference [16].
Acknowledgments
The authors thank Theophile Gaudin for useful discussions. A A-G acknowledges generous support from the Canada 150 Research Chair Program, Tata Steel, Anders G Froseth, and the Office of Naval Research. We acknowledge supercomputing support from SciNet. M K acknowledges support from the Austrian Science Fund (FWF) through the Erwin Schrödinger fellowship No. J4309. F H acknowledges support from the Herchel Smith Graduate Fellowship and the Jacques-Emile Dubois Student Dissertation Fellowship. P F has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Grant No. 795206.
Supplementary data (0.3 MB PDF)