Abstract
Brain organoid technology has transformed both basic and applied biomedical research and paved the way for novel insights into developmental processes and disease states of the human brain. While the use of brain organoids has been rapidly growing in the past decade, the accompanying bioengineering and biofabrication solutions have remained scarce. As a result, most brain organoid protocols still rely on commercially available tools and culturing platforms that had previously been established for different purposes, thus entailing suboptimal culturing conditions and excessive use of plasticware. To address these issues, we developed a 3D printing pipeline for the fabrication of tailor-made culturing platforms for fluidically connected but spatially separated brain organoid array culture. This all-in-one platform allows all culturing steps—from cellular aggregation, spheroid growth, hydrogel embedding, and organoid maturation—to be performed in a single well plate without the need for organoid manipulation or transfer. Importantly, the approach relies on accessible materials and widely available 3D printing equipment. Furthermore, the developed design principles are modular and highly customizable. As such, we believe that the presented technology can be easily adapted by other research groups and fuel further development of culturing tools and platforms for brain organoids and other 3D cellular systems.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 4.0 license. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
Brain organoids are three-dimensional (3D) neural aggregates self-assembled from differentiating pluripotent stem cells (PSCs) in suspension culture [1–3]. As such, they provide a unique human in vitro platform that more closely mimics the structural, functional, and cytoarchitectural aspects of the developing native brain than the traditional two-dimensional cell cultures [4–6]. Coupled with gene editing tools [7], multiomics analysis [8], electrophysiological investigation [9], or transplantation procedures [10, 11], organoid-based studies have yielded new important insights into evolutionary questions [12], developmental processes [13], and disease states [14] of the human brain.
While the use of brain organoids has been rapidly expanding in recent years, bioengineering and biofabrication solutions aimed at improving brain organoid protocols have been scarce even though the convergence of these technologies might play an important factor in moving the field forward [15, 16]. For instance, it has been shown that adding a biocompatible scaffold—secondary to the embedding hydrogel network—improves neuronal differentiation, organoid formation and long-term maintenance [16–20]. Customized platform engineering has also provided benefits for brain organoid culture. Polydimethylsiloxane (PDMS) soft lithography, a standard method for microfluidic system fabrication, has been leveraged to develop platforms for increased organoid viability [21] and automated organoid culturing [22]. PDMS chips have been further used to study brain organoid formation under exposure to nicotine [23], cannabis [24] and breast cancer derived exosomes [25]. Notably, recent advances in 3D printing technology have provided modular, customizable, low-cost, and easily accessible approaches for platform fabrication. For example, 3D printed miniaturized spinning bioreactors have reduced culturing media volume and incubator space in comparison to the commercial spinning bioreactors used for long-term maintenance of brain organoids [26, 27]. Also, custom-made 3D printed microfluidic systems allowed for optical monitoring during organoid development [28] and provided an environment for brain organoid vascularization [29]. However, most of these approaches are aimed at maintenance of already formed organoids in terminal stages of differentiation. Cellular aggregation and hydrogel embedding are still performed in commercial well plates leading to excessive use of expensive plasticware and need for physical handling of organoids between different protocol steps.
Here, we provide a simple pipeline for the fabrication of custom-made well plate platforms for brain organoid formation, hydrogel embedding, and maintenance. The fabrication methodology combines easily accessible 3D printing technology with PDMS soft lithography, without the need for access to cleanroom facilities or highly specialized equipment. The developed well plate design provides a modular all-in-one platform that replaces all the commercial plasticware used in brain organoid protocols and removes the potentially harmful organoid handling and transfer steps. Via immunocytochemistry and real-time quantitative polymerase chain reaction (RT-qPCR), we show that human ventral midbrain (vMB) organoids can be generated in our culturing platform on par with the conventional protocols. Low-cost of the process, fast prototyping capability, and material biocompatibility render this approach adaptable to organoid protocols and experimental needs beyond the ones demonstrated in our work.
2. Materials and methods
2.1. 3D printing
The soft lithography molds, the outer frame, and well insert with slits were designed with Fusion 360 software (Autodesk, Inc.) and exported as Standard Tesselation Language (.stl) files into the slicing software of the 3D printer (Preform Software V 2.18.0 Formlabs Inc). The soft lithography molds were printed with a commercial photopolymer (Black resin V4, Formlabs) in a Form 3 stereolithography (SLA) printer (Formlabs) operating with a source wavelength of 405 nm and layer thickness resolution of 25 μm. The biocompatible components were prepared with another commercial photopolymer (Dental clear LT V2, Formlabs) in a Form 2 SLA printer (Formlabs) operating with a source wavelength of 405 nm and layer thickness of 100 μm. After 3D printing, the components were carefully washed with fresh isopropanol three times and dried by nitrogen blowing. Then, the 3D printed components were post-cured at 60 °C for 60 min in a Form Cure chamber (Formlabs). Finally, the 3D printed components were immersed in fresh isopropanol overnight and dried at room temperature.
2.2. Soft lithography
The bottom element serving as a substrate for brain organoid formation and culturing was made from SYLGARD 184 PDMS (Dow, #1673921) using a standard soft lithography technique. Elastomer was mixed with the corresponding curing agent in a ratio of 10:1 and poured in the 3D printed mold. Degassing was performed in a plastic desiccator (Rotilabo, Carl Roth) for 20 min under applied vacuum. Elastomer was cured at 60 °C overnight and then peeled off from the mold and trimmed using a scalpel. Culturing wells were then coated with a thin layer of less viscous Sylgard 184 elastomer mixed with a curing agent in 5:1 ratio. The coating was performed with minimal amount of PDMS using a cotton swab. Curing of the coating was performed at 60 °C overnight. Fabricated PDMS parts were soaked in 70% ethanol overnight, rinsed twice in sterile filtered Milli-Q (Millipore) water, and immersed in fresh Milli-Q water overnight before drying under constant sterile laminar air flow in a tissue culture hood.
2.3. Human PSC (hPSC) culture
Undifferentiated RC17 (Roslin Cells, cat. No. hPSCreg RCe021-A) were maintained in iPS Brew medium (Miltenyi) in 6-well plates coated with 0.5 µg cm−2 Lam-521 (Biolamina, LN-521) and passaged using 0.5 mM EDTA.
2.4. vMB organoid culture
Human vMB organoids were generated by following previously published protocol [18, 30] in both conventional wells and the 3D printed platform. Single-cell suspension was generated by detaching hPSCs from the culturing dish using 0.5 mM Accutase (Thermo Fisher Scientific). 8000 cells in 200 µl iPS Brew medium supplemented with 10 μM Y-27632 dihydrochloride (Miltenyi, #130-106-538) were seeded in each 3D printed well. After 3 d (day 0), neural differentiation was initiated by replacing iPS Brew medium with 1:1 Dulbecco's Modified Eagle Medium (DMEM)/F12:Neurobasal (Thermo Fisher Scientific, #21331020 and #A1371201), 1:100 N2 supplement (Thermo Fisher Scientific, #A1370701), 10 μM SB431542 (Miltenyi, #130-106-543), 150 ng ml−1 rhNoggin (Miltenyi, #130-103-456), 400 ng ml−1 SHH-C24II (Miltenyi, #130-095-727), 1.5 μM CHIR99021 (Miltenyi, #130-106-539), 200 mM L-glutamine (Thermo Fisher Scientific, #25030081) and 10 000 U ml−1 penicillin-streptomycin (Thermo Fisher Scientific, #15140122). On day 11, the medium was replaced with 1:50 Neurobasal medium, B27 supplement without vitamin A (Thermo Fisher Scientific, #12587010) and 100 ng ml−1 FGF-8b (Miltenyi, #130-095-740). At day 14, vMB organoids were embedded in 30 µl droplets of Matrigel (BD Biosciences). From day 16 onward, 0.5 mM db-cAMP (Sigma-Aldrich, #D0627-1G) and 1 μM DAPT (R&D Systems, #2634) were added to medium for terminal maturation until the experimental endpoint. Differentiation medium was supplemented with 1% minimum essential medium nonessential amino acids (MEM-NEAA, Sigma-Aldrich, #M7145) and 0.1% 2-mercaptoethanol (Merck, #8057400005) throughout the entire differentiation period.
For the conventional protocol, the initial cell seeding was done in ultra-low attachment round bottom 96-wells (Corning, CLS7007). On day 6, organoids were transferred to a 24-well plate. Embedding at day 14 was performed inside PARAFILM (Sigma Aldrich, P7793) dimples as described before [31]. After embedding, vMB organoids were transferred in a low-attachment 6-well plate where they remained until the experimental endpoint.
2.5. Organoid cryosectioning and immunocytochemistry
Organoids were fixed in 4% paraformaldehyde (PFA, Merck Millipore, 1040051000) for 5 h at room temperature. They were then washed three times with DPBS (−Ca2 ± Mg2+, Thermo Fisher Scientific, #A1285601) and immersed in 30% w/v sucrose (Fisher Chemical. S/8600/60) overnight. Sucrose was then replaced with 1:1 mixture of OCT (HistoLab, 45830) and 30% sucrose incubated at 4 °C for 6 h on an orbital shaker. Organoids were then transferred in a cryomold, snap-frozen on dry ice, and placed in −80 °C for long term storage until cryostat sectioning (Fisher Scientific, CryoStar NX70). 20 µm cryosections were transferred on glass slides, washed three times with DPBS and postfixed with 4% PFA for 10 min at room temperature. Sections were incubated in 0.3% Triton X-100 and 5% donkey serum (Merck Millipore, S30) for at 1 h and left in primary antibody solution at 4 °C overnight. Sections were then washed three times with DPBS and incubated with secondary antibody solution for 1 h and mounted with polyvinyl alcohol mounting medium with DABCO (PVA-DABCO, Sigma Aldrich, 10981) containing DAPI (1:1000, Sigma Aldrich, D9542). Imaging was performed on an inverted laser scanning confocal fluorescence microscope (A1RHD, Nikon) equipped with a 20x objective. Images were analyzed in ImageJ (NIH).
2.6. Calcium imaging
Organoids were incubated for 30 min at 37 °C with 3 µM Calbryte 520 AM calcium indicator (AAT Bioquest) in BrainPhys Imaging Optimized Medium (STEMCELL Technologies) with 0.02% Pluronic F-127 (Sigma Aldrich). Organoids were then rinsed twice in BrainPhys. Imaging was performed at 37 °C and 5% CO2 on an inverted confocal microscope (ECLIPSE Ti2-E, Nikon) equipped with a spinning disk module (CSU-W1, Yokogawa). Images were acquired at 5 Hz for 2 min through a 40x water immersion objective (N.A. 1.25). Images were analyzed in ImageJ (NIH) and plotted in Prism (GraphPad).
2.7. Dopamine detection
HEK-293 Flp-In T-Rex cell line stably expressing fluorescent G protein-coupled receptor-based DA sensor GRABDA2M [32, 33] were cultured in DMEM supplemented with 10% FBS, 15 µg ml−1 blasticidin, and 200 µg ml−1 Hygromycin B. 24 h before the measurements were taken, sniffer cells were seeded in imaging chambers (18-well, ibidi) coated with poly-L-ornithine and the expression of GRABDA2M sensor was induced with 1 µg ml−1 tetracycline. Live DA sniffer cell imaging was performed on a widefield Leica microscope using a 20x NA 1.4 objective. Each measurement on the graph corresponds to 80 µl HBSS in which 3 organoids were incubated for 1 h. After collection, samples were snap frozen and kept at −80 °C until measurements were performed. Images were aligned and quantified in ImageJ (NIH) and results plotted in Prism (GraphPad).
2.8. RT-qPCR
The vMB organoids (three per sample) were washed two times with DPBS for 10 min. The samples were then lysed, snap-frozen, and total RNA was collected using the RNeasy Micro Kit (QIAGEN#74004) according to the instructions provided by the manufacturer. 1 µg of RNA was reverse transcribed to cDNA with the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher #K1642). Generated cDNA, primers of interest and SYBR Green Master mix (Roche #04887352001) were mixed using the Bravo automated liquid handling platform (Agilent) and analyzed with a LightCycler 480 II instrument (Roche) via a two-step protocol with a 95 °C, 30 s denaturation step followed by a 60 °C, 60 s annealing/elongation step for a total of 40 cycles. Results were analyzed in Excel and plotted in Prism (GraphPad). Statistical differences between conditions was assessed by Mann–Whitney test.
2.9. Scanning electron microscopy
PDMS samples were mounted on 25 mm aluminum stubs, sputter coated with 10 nm Platinum/Palladium (80:20) and imaged in a Jeol JSM-7800F scanning electron microscope.
3. Results
3.1. 3D printed platform design and assembly
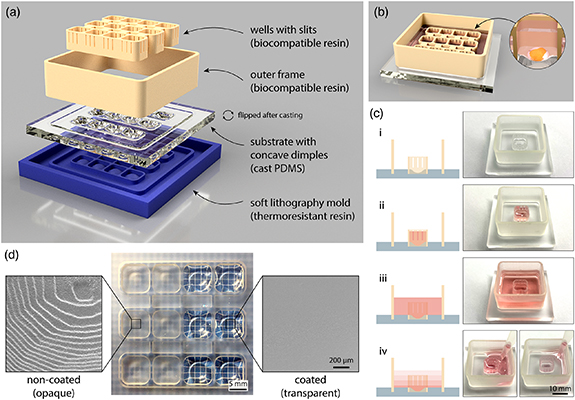
The cell culturing platform developed here consists of three main components: a PDMS substrate with concave dimples, an insert with 3D printed wells with open slits, and a 3D printed outer frame (figure 1(a)). The 3D printed elements click into the grooves designed in the substrate to allow quick and easy assembly of the 3D printed platform for brain organoid generation and maintenance. Since all the three elements including the mold for PDMS casting, are 3D printed via SLA, the design process is fully customizable and compatible with fast prototyping workflows. Here, we designed a 3D printed platform with 12 individual wells. Each well is rectangular in shape with 8 mm sides and 6 mm high walls. Concave dimples are 1.8 mm deep at the lowest point. As such, each well is designed to fit a single human brain organoid (figure 1(b)).
Figure 1. Schematic overview of assembly and function of the 3D printed platform with fluidically connected wells for brain organoid formation and maintenance. (a) 3D rendering of different elements that are required to prepare the 3D printed platform. PDMS substrate is flipped around after casting. (b) 3D rendering of the assembled platform (PDMS cast, outer frame, and well insert) containing 12 fluidically connected wells designed to accommodate individual brain organoids (zoomed-in image). (c) Illustration (left) and photographs (right) showing different modes of liquid handling in the platform: (i) empty platform; (ii) medium contained and exchanged only inside the well; (iii) medium contained both inside and around the well; (iv) fluidic connection between the well and the surrounding space via the slits allows medium exchange throughout the platform by pipetting at a single point outside of the well. For illustration, liquid handling was demonstrated in a single-well platform. (d) Photograph showing the PDMS cast substrate with dimples before (left) and after (right) coating with a thin layer of PDMS. Electron micrographs show surface roughness of non-coated (opaque) and coated (transparent) dimple surface at the microscale.
Download figure:
Standard image High-resolution imageKey features in the 3D printed platform are the slits on the sides of the wells. These slits are 400 µm wide and there are 3 slits on each side of the well. As such, open slits serve multiple functions (figure 1(c)). Hydrophobicity of the material and the specific slit dimensions allow for the cell culture medium to fill the wells without leaking through the slits into the surrounding compartment. Furthermore, the dimension of the slits prevents brain organoids—at any stage of differentiation—to flow out of their designated wells. Wider slits would not be able to stop cellular aggregates of only several hundred micrometers in size from being pushed out of the wells by liquid flow during medium changes. Finally, when the outer areas of the 3D printed platform are filled with medium and wells become wet from both sides, the slits no longer present a liquid barrier thus resulting in fluidic connection of the whole platform. This allows for simultaneous single-step medium exchanges in all wells through pipetting in one corner of the 3D printed platform without direct disturbance of individual brain organoids.
Non-invasive morphological tracking of brain organoid formation is important for assessment of the experimental progress. This requires the concave dimples in the PDMS substrate to be transparent in order to allow for optical microscopy through the bottom of the 3D printed platform. Furthermore, the surface of the dimples has to be smooth to facilitate reproducible cellular aggregation and self-assembly. However, casting PDMS on 3D printed molds leads to a rough and opaque surface due to replication of individual 3D printed layers visible at the microscale using scanning electron microscopy (SEM) (figure 1(d) left). Postprocessing of the cast parts by additional coating of the dimples with a thin layer of PDMS removes surface roughness and renders the surface of the dimples in the platform transparent and smooth (figure 1(d) right).
3.2. Brain organoid workflow in 3D printed well plates
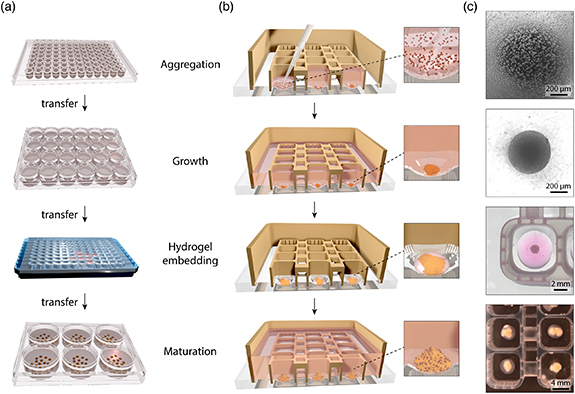
The conventional approach for the generation of brain organoids requires the use of at least four different types of well plates. During this procedure, cell aggregates and brain organoids are manually handled using a pipette when they are transferred to a new well plate, a process which might mechanically damage cellular structures or affect their differentiation and functionality (figure 2(a)). Furthermore, each organoid has to be transferred individually from one well to another, which is time consuming and might decrease reproducibility in experimental studies. In conventional protocols for brain organoid differentiation, the starting hPSCs are commonly aggregated in concave 96-well plates or microwells with low-attachment surfaces. Aggregates of differentiating and proliferating neuronal progenitors expand to a point where they consume more medium than what is available in that well plate format, and therefore need to be transferred to larger wells. Around day 14, brain organoids are positioned into dimples for embedding in hydrogel drops. Once the hydrogel has solidified, organoids are transferred in a six-well plate or a petri dish for terminal differentiation and maturation. In all these well plates, media exchange during culturing has to be done individually in each separate well.
Figure 2. Comparison of workflow for brain organoid formation using the conventional approach and 3D printed platform with fluidically connected wells. (a) Schematic overview of the different well-plate formats needed for brain organoid formation with conventional approach and commercial plates. In the descending order: ultra-low attachment U-bottom 96-well plate, 24-well plate, handmade parafilm dimples and 6-well plate. (b) Schematic overview of brain organoid formation in the 3D printed platform where organoids are not mechanically disturbed and transfer is not required. Magnified images show organoid formation progress in a single well. (c) Brightfield images (top two) and photographs (bottom two) of cellular structures at different timepoints during brain organoid formation inside fluidically connected 3D printed wells.
Download figure:
Standard image High-resolution imageIn the 3D printed platform, on the other hand, all the above-mentioned steps are performed in a single well plate avoiding transfer of cells after initial seeding (figure 2(b)). Single-cell suspension of hPSCs is added to each empty well where cells quickly settle at the bottom and aggregate into an embryoid body. hPSCs then expand and start differentiating towards vMB neuronal identity. When the volume of individual wells becomes insufficient to match the cellular consumption of growing brain organoids, increased media volume is supplied to cells by fluidically connecting all the wells in the well plate and adding media in a single-step process to the whole well plate. Notably, the 3D printed well plate allows hydrogel embedding of brain organoids directly within the same wells. For this purpose, medium is removed and a drop of hydrogel precursor is added directly to each 3D printed well. Following hydrogel solidification, the medium is again added to the plate and the medium volume is increased as needed during organoid growth and maturation.
3.3. Characterization of human vMB organoid formation
Since brain organoids are kept in the same wells with transparent bottoms throughout the entire differentiation protocol, the 3D printed plate platform enables monitoring of organoid formation via an inverted brightfield microscope (or a fluorescent microscope in case of reporter stem cell lines). hPSCs seeded into 3D printed well plates successfully aggregated into compact embryoid bodies with smooth edges that continued to grow in size overtime until the experimental endpoint (figure 2(c)). Importantly, we did not observe any significant cell death indicating high biocompatibility of the materials used for plate production and assembly. Live/dead staining at day 5 revealed negligible number of dead cells in the formed spheroids (figure 3(a)). At day 16, defined neuroectoderm layer formation was observed (figure 3(b)). At the same experimental timepoint, we performed immuoncytochemistry to assess whether the presence of photopolymerized resin and PDMS affected the patterning of neural vMB progenitors. The correct developmental identity was confirmed by the presence of vMB floorplate progenitor markers LMX1A, FOXA2, and OTX2 (figure 3(c)). Co-expression of these markers at day 16 is in line with previous work on vMB organoids [18, 30] and the reference 2D protocol for the generation of vMB dopamine (DA) neurons based on dual-SMAD inhibition and vMB patterning with SHH and GSK3i, followed by vMB progenitor caudalization with FGF8b [34, 35]. Correct vMB patterning was further verified by RT-qPCR (figure S1a). Self-organization of differentiating progenitors into neural rosettes was also observed (figure 3(d)). Immunostainings of cryosectioned vMB organoids cultured for 60 d in 3D printed platform revealed that the 3D cultures were rich in neurons (MAP2, TUJ1) that also displayed DA identity, marked by tyrosine hydroxylase (TH), on par with the control organoids generated via conventional protocol (figure 3(e)). Importantly, DA neurons were present throughout the brain organoid as can be seen by whole section imaging (figure 3(f)). Immunocytochemistry for apoptosis marker cleaved caspase-3 (CAS3) revealed that 3D printed well plates did not lead to increased apoptosis in brain organoids (figure 3(g)). RT-qPCRs indicated an overall tendency of higher expression of the pan-neuronal gene MAP2 and genes related to ventral midbrain identity (LMX1A, CORIN, FOXA2) as well as a significant increase in expression for genes associated to DA fate (NURR1, DDC, TH) compared to conventional plates (figure 3(h)). Further gene expression analysis (figure S1b) revealed presence of glial cells (PDGFRA, GFAP), vascular and leptomeningeal cells (COL1A1, LUM), cortical glutamatergic neurons (EMX1, vGLUT), GABAeric neurons (GAD2), interneurons (VGAT, SST, CHAT, SERT), and medium spiny neurons (DARPP32, CTIP2). Live calcium imaging experiment confirmed that neurons within the generated vMB organoids fire spontaneous action potentials, a sign of a healthy neuronal population (figure 3(i), movie S1). Furthermore, we showed that cultured neurons are able to release dopamine (∼3 nM per organoid), measured using GRABDA2M sniffer cells (figure 3(j)). We have also verified that the 3D printed platform is suitable for the generation of non-embedded brain organoids. In conventional protocols, multiple non-embedded brain organoids are commonly transferred into the same well to avoid the slow handling of large numbers of samples, which frequently leads to organoid fusion (figure S2(a)). Fused organoids can be mechanically separated but always at the expense of tissue damage. Non-embedded organoids could still fuse in orbital shaker under constant stirring. On the contrary, our platform keeps the organoids in spatially separated positions to prevent fusion while allowing easy medium changes. In line with what was observed in the embedded brain organoids, immunostained cryosections of non-embedded vMB organoids at day 60 were also rich in cells expressing the pan-neuronal markers MAP2 and TUJ1 and further contained TH-expressing neurons (figure S2(b)). Accordingly, gene expression analysis showed an increase in genes related to mature neurons, DA neuronal fate, as well as vMB identity (figure S2(c)).
Figure 3. Analysis of vMB organoid formation and function in the 3D printed platform. (a) Confocal fluorescence image displaying live (Calcein AM) and dead (EthD-1) cells in a spheroid 5 d after cell seeding. (b) Microscope brightfield image of a neural progenitor aggregate at day 16. (c) Confocal fluorescence image of progenitor markers in a spheroid at day 16. (d) Confocal fluorescence image of a neural rosette formation. (e) Confocal fluorescence image of pan-neuronal (MAP2, TUJ1) and DA (TH) markers in cryosectioned brain organoids at day 60. (f) Immunocytochemistry of whole vMB organoid cryosection at day 60. (g) Immunocytochemistry of cleaved caspase-3 (CAS3) and quantification of the marker over DAPI. (h) RT-qPCR gene expression analysis for neuronal and dopaminergic genes in vMB organoids at D60 generated in commercial well plates and in 3D printed platform. n = 3 (i) selected image from an acquired time series displaying the neuronal network and regions of interest (ROIs) analyzed for changes in fluorescence intensity (left). Differential fluorescence intensity profiles showing changes in intracellular Ca2+ as a function of time (right). (j) Live fluorescence images showing response of GRABDA2M sniffer cells to increased concentration of dopamine (top) and generated response curve of the same cells (bottom left). Quantification of dopamine from samples collected from day 60 vMB organoids cultured in control and 3D printed well plates (bottom right).
Download figure:
Standard image High-resolution image4. Discussion and conclusion
The 3D printed platform presented here provides a method for simultaneous culturing of multiple brain organoids in a fluidically interconnected wells within the same plate. Control over fluidic connection of the wells allows single-step medium change for the whole well plate without mechanical perturbation of sensitive 3D cultures. Since the fabrication of the platform relies fully on a widely available 3D printing methodology, it can be used for fast prototyping of new designs that can accommodate different types of 3D cultures or high throughput applications (e.g. creating a fluidically interconnected 96-well plate). Furthermore, curvature and depth of the concave dimples can easily be altered to change cell aggregation dynamics [36]. Clear bottom of the wells allows for optical monitoring of organoid progress which could provide a method for non-invasive longitudinal phenotyping of organoid formation and growth in chemical screens [37, 38]. Our data further provides evidence that biocompatible commercial resins, such as the resin used here for printing of culture wells and platform walls, if adequately postprocessed, do not interfere with stem cell differentiation and patterning [39]. This allows for the development of novel 3D printed devices for neuronal in vitro applications. Our platform could also be placed onto an orbital shaker to facilitate long-term maturation of brain organoids and minimize the effects of low oxygen and nutrient supply to the core of the 3D cellular constructs [31]. Furthermore, 3D printed plates could be integrated into a culturing system with continuous media flow as an alternative solution for improved cell fitness [21]. Finally, our platform could also be used to fuse together different region-specific brain organoids to form assembloids, as models of inter-regional neuronal connectivity [40, 41].
Acknowledgments
The authors would like to thank Bengt Mattsson for the creation of 3D models and illustrations. Lund University Bioimaging Centre (LBIC) is gratefully acknowledged for providing experimental resources for electron and fluorescence microscopy. B R and S S K acknowledge the financial support from the European Research Council under the Horizon 2020 framework program (Grant No. 772370-PHOENEEX). J K and M P acknowledge the financial support from The Swedish Research Council (2021-02967) and HORIZON-EIC-2021-PATHFINDEROPEN-01 project OpenMIND (101047177). A F is supported by Swedish Research Council (2022-01432) and Italian Ministry of Health (B53C22007860001).
Data availability statement
All data that support the findings of this study are included within the article (and any supplementary files).
Supplementary data (0.8 MB PDF)
Supplementary video 1 (30.1 MB AVI)