Abstract
Biomaterial design in tissue engineering aims to identify appropriate cellular microenvironments in which cells can grow and guide new tissue formation. Despite the large diversity of synthetic polymers available for regenerative medicine, most of them fail to fully match the functional properties of their native counterparts. In contrast, the few biological alternatives employed as biomaterials lack the versatility that chemical synthesis can offer. Herein, we studied the HUVEC adhesion and proliferation properties of elastin-like recombinamers (ELRs) that were covalently functionalized with each three high-affinity and selectivity αvβ3- and α5β1-binding bicyclic RGD peptides. Next to the bicycles, ELRs were also functionalized with various integrin-binding benchmark peptides, i.e. knottin-RGD, cyclo-[KRGDf] and GRGDS, allowing for better classification of the obtained results. Covalent functionalization with the RGD peptides, as validated by MALDI-TOF analysis, guarantees flexibility and minimal steric hindrance for interactions with cellular integrins. In addition to the covalently modified RGD-ELRs, we also synthesized another benchmark ELR comprising RGD as part of the backbone. HUVEC adhesion and proliferation analysis using the PicoGreen® assay revealed a higher short-term adhesion and proliferative capacity of cells on ELR surfaces functionalized with high affinity, integrin-binding bicyclic RGD-peptides compared with the ELRs containing RGD in the backbone.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
Introduction
A central goal of research in tissue engineering and regenerative medicine is the design of biomaterials that can be used to control critical aspects of cellular behavior. Such materials should guide cells toward the phenotypes and architectures that are needed to restore tissue function or induce cells from surrounding tissue to infiltrate implanted matrices [1], including vascular grafts [2], bone grafts [3], wound dressings [4], and injectable drug depots [5]. One major problem that still remains is the inadequate interaction between polymer and cells, which leads to foreign body reactions such as inflammation, infections, and implant encapsulation, as well as thrombosis and embolization, in vivo. Research into the surface modification of polymeric materials to guide cellular activity in biomaterials [6–8] designed for tissue-engineering applications has mostly focused on the use of natural extracellular matrix proteins and short peptides such as the integrin-binding tripeptide RGD [9, 10]. The use of small peptides offers several advantages, such as straightforward synthesis and a low immunogenic potential [11, 12]. Moreover, they tend to exhibit high stability towards sterilization conditions, heat treatment, pH-variation and storage [13, 14]. However, it is important to take into account that the peptide stability is strongly dependent on the sequence. In particular, peptides containing methionine, cysteine or tryptophan are prone to oxidation with resultant loss of bioactivity [15].
While a large variety of modified synthetic polymers have been extensively explored in regenerative medicine, only a few biological materials, mostly purified proteins from animal tissues, have been studied in detail to date [16]. In this regard, recombinant proteins have recently received interest as an attractive alternative for tissue-engineering applications and surface functionalization [17, 18]. Elastin-like recombinamers (ELRs) are some of the most intensively studied groups of recombinant proteins over the past decade [19]. ELRs comprise repetitive sequences comprising the pentapeptide 'Val-Pro-Gly-X-Gly', where the guest residue 'X' can be any amino acid except proline. These polypeptides are highly flexible due to weak hydrophobic interactions and hydrogen bonds that enable the chains to extend and retract similarly to a spring [20–22]. ELRs can be codified in synthetic genes and expressed in Escherichia coli in large quantities, thus allowing greater control of the amino acid sequence and molecular weight when compared with the chemical synthesis of large polypeptides [18]. Furthermore, due to their inverse temperature transition property, they can easily be separated from the raw protein mixture [23], thereby resulting in a purified, biocompatible, biodegradable, and non-immunogenic engineered protein [24]. ELRs have several advantages as regards tissue engineering applications, such as their defined macromolecular structure, controlled swelling behavior and porosity, degradability, and controllable mechanical properties [25, 26].
First described in 1992 by Nicol et al [27], ELRs comprising integrin-binding motifs such as RGD [1, 28–30] or REDV [30, 31] have been investigated as possible materials for biomaterial applications. However, thus far, recombinant synthesis has limitations in the incorporation of non-canonical amino acids and does not allow the formation of cyclized peptides [32]. Despite this, ELR hydrogels covalently functionalized with a peptide mimic of the receptor-binding region of VEGF (vascular endothelial growth factor) showed enhanced HUVEC proliferation over non-functionalized hydrogels [33].
One drawback of the non-integrin-selective RGD sequence is the inability to elicit responses based on closely defined intracellular pathways [34]. An elegant approach to circumvent this limitation, and an additional lever for the control of cell adhesion and proliferation, is orthogonal post-functionalization with peptides via modified lysine side-chains, for example, via maleimide-thiol coupling [34]. Functionalization via biorthogonal chemistry is a well-known strategy for the manufacturing of hydrogels and multifaceted cell culture scaffolds [35, 36]. Another possibility to functionalize biomaterials with peptides is via copper-free click chemistry [37]. For instance, Krause et al applied a metal free click reaction to functionalize alginate with a cyclic RGD peptide [38].
The integrin-binding RGD motif is generally applied in either linear or cyclic format, with the cell adhesion-promoting properties of these peptides depending on the material applied. Linear RGD derivatives, such as GRGDS, are still the most widely applied cell-adhesion sequences despite the fact that cyclic RGD peptides or RGD peptidomimetics provide much higher integrin affinities and selectivities [39, 40]. For example, ELRs functionalized with cyclo-[KRGDf] showed a 100% improved mouse osteoblast adhesion in comparison with ELRs functionalized with linear FGRGDS [16]. However, this study focused on the chemical functionalization of ELRs and did not include recombinant synthesized RGD-ELRs as a benchmark. In contrast, spider silk proteins genetically fused to GRGDSPG showed similar fibroblast adhesion and proliferation properties compared to spider silk proteins covalently modified with cyclo-[KRGDf] [41].
Recently, Bernhagen and coworkers reported short bicyclic RGD-peptides that bind to either integrin αvβ3, or to both integrins α5β1 and αvβ3, with high affinity [42]. For example, for integrin α5β1, the bicyclic peptide CT3RGDCT3AYJCT3 (J: D-Leucine, CT3 represents cysteines that were constrained using the trivalent scaffold 1,3,5-tris(bromomethyl)benzene) exhibited much higher inhibition (IC50: 90 nM) compared with monocyclic RGD-peptide cyclo-KRGDf or linear GRGDS (each IC50: >10 μM) [43]. Hence, bicyclic RGD-peptides potentially represent a new group of ligands that boost cellular adhesion and proliferation in biomaterials more than commonly applied linear RGD peptides.
In this study, we have created RGD-functionalized ELRs by conjugating various high-affinity integrin-binding bicyclic RGD-peptides to ELRs using copper-free click chemistry [44]. We then evaluated the cell adhesion and proliferation properties of peptide-ELR conjugates for HUVECs. ELRs functionalized with high-affinity bicyclic peptides were compared with ELRs functionalized with benchmark RGD peptides and ELRs that comprised RGD as part of their backbone at the short, medium and long term.
Materials and methods
ELR biosynthesis, modification and characterization
The ELRs used in this work were obtained using standard genetic engineering techniques [31]. They were purified using several cycles of temperature-dependent reversible precipitations by centrifugation below and above their transition temperature (Tt), thus making use of the intrinsic thermal behavior of these compounds [31]. The ELRs were subsequently dialyzed against purified water and freeze-dried. Two different ELRs, namely VKVx24, a structural recombinamer lacking a bioactive sequence, and HRGD6, a recombinamer containing the universal cell adhesion epitope (RGD) repeated six times per ELR molecule, were obtained. The purity and chemical characterization of these ELRs were verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) amino acid composition analysis, differential scanning calorimetry (DSC) and nuclear magnetic resonance (NMR) spectroscopy [31, 45]. ELRs were chemically modified by transformation of the ε-amine group in the lateral lysine chain to bear azide groups [46, 47]. VKVx24-N3 and HRGD6-N3 were prepared and characterized by NMR, Fourier transform infrared spectroscopy (FTIR), and DSC (supporting information is available online at stacks.iop.org/BMM/14/035009/mmedia).
Reagents and chemicals
Incubation and washing buffers were prepared using standard protocols. Amino acids were purchased from Iris Biotech (Marktredwitz, Germany) and Matrix Innovation (Quebec, Canada). Resins were purchased from Rapp Polymere (Tübingen, Germany) and Merck (Darmstadt, Germany). 1,3,5-Tris(bromomethyl) benzene (T3) and (1R,8S,9S)-Bicyclo[6.1.0]non-4-yn-9-ylmethyl N-succinimidyl carbonate (BCN-NHS) were purchased from Sigma-Aldrich (Steinheim, Germany). 2-Azidoethyl (2,5-dioxopyrrolidin-1-yl) carbonate was purchased from GalChimia (A Coruña, Spain).
Peptide synthesis
Linear peptide (3c) and linear precursor peptides of (1a–c), (2a–2c), (3a) and (3b) (figure 1) were synthesized using a fully automated peptide synthesizer from Gyros Protein Technologies (Symphony) by Fmoc-based solid-phase peptide synthesis on Rink-amide resin using standard coupling protocols. Folding of knottin-RGD peptide (3a) and backbone cyclization of cyclic RGD peptide (3b) were performed according to previously published protocols [48]. For the formation of bicyclic peptides (1a–c, 2a–c), purified linear peptides were dissolved at 0.5 mM in 1:3 MeCN/H2O, and 1.1 equiv. 1, 3, 5-tris(bromomethyl) benzene (T3) dissolved in MeCN (10 mM) and 1.4 equiv. ammonium carbonate (0.2 M in H2O) were added. After completion (30–60 min, monitored by UPLC/MS), the reaction was quenched with 10% TFA/H2O to pH < 4, followed by lyophilization. All peptides were purified by preparative HPLC on an RP-C18 column (Reprosil-Pur 120 C18-AQ 150 × 20 mm, Dr Maisch GmbH, Ammerbuch, Germany) using a MeCN/milliQ gradient (5%–65%) including 0.05% TFA followed by lyophilization (Christ Alpha 2–4 LDplus). An overview of all peptides can be found in table 1.
Figure 1. Overview of RGD-peptide–cyclooctyne conjugates for the functionalization of ELRs. (1a): K(BCN)-linker-CT3HPQcT3RGDcT3; (1b): K(BCN)-linker-CT3HPQCT3RGDcT3; (1c): K(BCN)-linker-CT3HSQCT3RGDcT3; (2a): K(BCN)-linker-CT3RGDcT3AYJCT3 (J: D-Leu); (2b): K(BCN)-linker-CT3RGDcT3AWGCT3; (2c): K(BCN)-linker-CT3RGDcT3AYaCT3; (3a): K(BCN)-linker-GCS-SRPRPRGDNPPLTCS-SSQDSDCS-SLAGCS-SVCS-SGPNGFCS-SG (K(BCN)-linker-knottin-RGD); (3b): cyclo-[K[K(BCN)-linker]RGDf]; (3c): K(BCN)-linker-GRGDS. Abz: 4-aminobenzoic acid.
Download figure:
Standard image High-resolution imageTable 1. Overview of ELRs investigated in this study, including degree of functionalization, RGD peptide and integrin selectivity. ELR labels containing 'P0' serve as controls that were not covalently modified via copper-free click reaction.
| ELR reference | Azide function-alization [%] | ELR modification or coating | Integrin selectivity |
|---|---|---|---|
| P1a | 5 | K(BCN)-linker-CT3HPQcT3RGDcT3 | αvβ3a |
| P1a-10 | 10 | ||
| P1b | 5 | K(BCN)-linker-CT3HPQCT3RGDcT3 | αvβ3a |
| P1b-10 | 10 | ||
| P1c | 5 | K(BCN)-linker-CT3HSQCT3RGDcT3 | αvβ3a |
| P1c-10 | 10 | ||
| P2a | 5 | K(BCN)-linker-CT3RGDcT3AYJCT3 | α5β1, (αvβ3)a |
| P2a-10 | 10 | ||
| P2b | 5 | K(BCN)-linker-CT3RGDcT3AWGCT3 | α5β1, (αvβ3)a |
| P2b-10 | 10 | ||
| P2c | 5 | K(BCN)-linker-CT3RGDcT3AYaCT3 | α5β1, (αvβ3)a |
| P2c-10 | 10 | ||
| P3a | 5 | K(BCN)-linker-GCoxPRPRGDNPPLTCox | αvβ3, |
| P3a-10 | 10 | QDSDCoxLAGCoxVCoxGPNGFCoxG | α5β1, αvβ5a |
| P3b | 5 | cyclo-[DfK(K(BCN)-linker)RG] | αvβ3, αvβ5a |
| P3b-10 | 10 | ||
| P3c | 5 | K(BCN)-linker-GRGDS | multiplea |
| P3c-10 | 10 | ||
| P0-RGD | — | RGD as part of the ELR-backbone | multiple |
| P0 | — | — | — |
| P0-FN | — | Fibronectin | αvβ3, α5β1 |
| P0-BSA | — | Bovine serum albumin (BSA) | — |
| J: D-Leu |
aData from competition ELISA [43].
Synthesis of peptide-cyclooctyne conjugates
BCN-NHS ester (1.1 equiv.) and 10 equiv. N, N-diisopropylethylamine were added to the peptides dissolved in DMSO (5 mM, TEC218 10 mM). After completion of the reaction (15–30 min, monitored by UPLC/MS), the reaction was quenched with 10% TFA/DMSO to pH < 4. The product was directly purified by preparative HPLC using a MeCN/milliQ gradient (5%–65%) including 0.05% TFA, followed by lyophilization. Conjugation was verified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectroscopy using a Voyager STR apparatus from Applied Biosystems.
Formation of peptide-functionalized ELRs
The ELRs comprising 5% or 10% peptide functionalization were modified based on the following calculations: azide-functionalized ELRs (62 809 Da) comprised 24 lysine molecules, 80% of which were functionalized with azides (NMR), corresponding to 19.2 azide groups per ELR molecule. The application of 1 equiv. BCN-functionalized peptide would result in the functionalization of 5.21% of the azide groups. Hence, application of 0.96/1.92 equiv. BCN-functionalized peptide would result in 5%/10% functionalization. As such, the application of 3 equiv. and 4 equiv. BCN-functionalized peptide would result in 15% and 20% functionalization. The conjugation of ELRs with BCN-functionalized peptides was performed as follows: azide-functionalized ELRs were dissolved in milliQ water (4 °C, 20 mg ml−1). BCN-functionalized peptides were dissolved at 5 mM in milliQ water (2a) in 50% MeCN/milliQ) and added to the ELR solutions. After shaking the copper-free click reactions for at least 24 h at 4 °C, the products were freeze-dried. Conjugation was subsequently verified by MALDI-TOF MS.
Analysis of turbidity by UV/vis-spectroscopy
The ELRs comprising 5% or 10% peptide functionalization were dissolved in milliQ water (4°C, 1 mg ml−1), and turbidity measurements were performed at a wavelength of 350 nm (Agilent Technologies Cary Series UV/vis Spectrophotometer). The transition temperature (Tt) of each RGD peptide-functionalized ELR (ELR-Peptide) was detected by performing a temperature ramp analysis.
Adsorption of peptide-functionalized ELRs on TCPS
The ELRs comprising 5% or 10% peptide functionalization were dissolved at 1 mg ml−1 in DPBS (Dulbecco's Phosphate Buffered Saline) –Ca–Mg (O/N at 4 °C) then added to a 96-well tissue culture polystyrene (TCPS) plate for incubation with ELR-Peptide solution O/N at 4 °C with gentle shaking. A positive control was obtained by incubating fibronectin at 10 μg ml−1 in DPBS –Ca–Mg, whereas a negative control was obtained by incubating BSA at 5 mg ml−1 in DPBS –Ca–Mg. The plates were subsequently sterilized using an ultraviolet (UV) lamp for 20 min, the solutions removed and the wells washed twice with DPBS –Ca–Mg. Plates were then incubated with a 5 mg ml−1 BSA blocking solution at 37 °C for 2 h. Finally, the solution was removed and the plates washed twice with DPBS –Ca–Mg prior to cell seeding.
Contact angle measurements
Contact angle (CA) measurements were performed using the sessile drop method on a Data Physics OCA20 System instrument. The drop profile images during micro-syringe dispensation were recorded using an adapted CCD video camera. Measurements were taken at ambient temperature 5 s after application of the drops to the surfaces. The CA values are the average of ten measurements, including standard deviation, for different locations on each surface in order to ensure a representative value of the contact angle.
X-ray photoelectron spectroscopy (XPS)
XPS experiments were carried out using a Physical Electronics (PHI) 5500 spectrometer equipped with a monochromatic x-ray source (Al Ka line, energy: 1486.6 eV and 350 W). The pressure inside the analysis chamber was 10−7 Pa. All measurements were performed at an angle of 45° with respect to both the x-ray source and analyzer. Survey scans were taken in the range 0–1100 eV, with a beam diameter of 200 μm, and high-resolution scans were obtained for C1s, N1s, O1s. The elemental surface composition was estimated from the area of the different photoemission peaks taken from the survey scans modified by their corresponding sensitivity factors.
Cell culture and cell adhesion assay
Human umbilical vein endothelial cells (HUVECs) (Cat.# C-015-10C; Gibco) at passage 2 were used in all experiments. HUVECs were cultured in Medium 200 (Gibco) supplemented with Low Serum Growth Supplement (LSGS) kit (Gibco), thus resulting in the following final concentrations: fetal bovine serum (FBS) 2% (v/v); hydrocortisone 1 μg ml−1; human epidermal growth factor 10 ng ml−1; basic fibroblast growth factor 10 ng ml−1 and heparin 10 μg ml−1. A separate vial of Gentamicin/Amphotericin solution (Gibco) was also incorporated at a final concentration of 10 and 0.25 μg ml−1, respectively. HUVECs were incubated at 37 °C and 5% CO2 and harvested at 90% confluence by trypsin–EDTA treatment.
HUVECs were seeded at a density of 5300 cells cm−2 in serum-free Medium 200 (Gibco) for 30 min on different surfaces (n = 3), and allowed to adhere for 30 min, after which time Medium 200 was removed and the cells cultured in LSGS-supplemented Medium 200 for 14 d. HUVEC adhesion and spreading were evaluated after incubation for 30 min, 4 h and 1, 3, 5, 7 and 14 d. Cultures were provided with fresh media daily, up to the time of the staining period or DNA quantification.
DNA analysis
DNA content was determined using the PicoGreen® assay after incubation for 4 h and 1, 3, 5, 7 and 14 d. Briefly, the cells were lysed with a solution of 0.1% Triton X-100 (Sigma Aldrich) in phosphate buffered saline (PBS) (v/v) [49], and the PicoGreen® analysis for DNA content was performed in 96-well plates at standard fluorescein wavelengths (excitation at 480 nm and emission at 520 nm) according to the manufacturer's instructions (Invitrogen) using an automated plate reader (Bionova Cientifica, Molecular Devices). Fluorescence intensities were transformed into cell numbers using a calibration curve obtained by measuring the fluorescence of defined cell amounts (n = 0, 100, 1000, 10 000 and 100 000):

2D immunofluorescent staining
Immunofluorescent staining of actin (red), vinculin (green), and nuclei (blue) was performed to visualize the HUVECs on different surfaces. After cell culture, cells were fixed with 4% (w/v) paraformaldehyde, permeabilized with 0.1% Triton-X 100 and blocked with 1% bovine serum albumin (BSA) in PBS solution. Focal contact formation was evaluated by incubating overnight at 4 °C antivinculin rabbit monoclonal antibody (AlexaFluor® 488) (1:200) (Abcam). Cell actin cytoskeletons and nuclei were stained with Rhodamin phalloidin (1:80) (Invitrogen) and DAPI (1:10000) (Lonza), respectively. Cell adhesion and morphological changes were examined using an inverted fluorescence microscope (Nikon Eclipse Ti E) and its associated software. A scan of each surface at low magnification (10×) was analyzed for this study. The images are representative of the morphology found in the samples studied, with at least two to three captures per well being taken. Quantitative image analysis of cell spreading area has been performed at short time incubation (4 h); the cell area was calculated by the software associated with the microscope Nikon Eclipse Ti E.
Statistical analysis
Values are expressed as mean ± standard deviation. Data were analyzed by performing the normality test Shapiro0–Wilk. The parametric data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey's Honestly Significant Difference post hoc test; if only two groups were being compared, an unpaired t-test was used instead of ANOVA to assess the statistical difference. The nonparametric data were analyzed performing the Kruskal–Wallis test followed by Dunn's multiple-comparison test. All statistical analyzes were performed with GraphPad Prism. A P-value lower than 0.05 was considered to be statistically significant.
Results
Selection and synthesis of RGD peptides
For this work, we selected three high-affinity integrin αvβ3- and α5β1/αvβ3-binding bicyclic peptides for the synthesis of peptide-cyclooctyne conjugates (figure 1). The αvβ3-binding peptides are K(BCN)-linker-CT3HPQcT3RGDcT3 (1a), K(BCN)-linker-CT3HPQCT3RGDcT3 (1b), and K(BCN)-linker-CT3HSQCT3RGDcT3 (1c), whereas the α5β1/αvβ3-binding peptides are K(BCN)-linker-CT3RGDcT3AYJCT3 (2a), K(BCN)-linker-CT3RGDcT3AWGCT3 (2b), and K(BCN)-linker-CT3RGDcT3AYaCT3 (2c). The linker sequence PPPSG-(Abz)-SG was designed based on the HexPPP spacer reported by Pallarola et al [47] 'T3' stands for the scaffold derived from 1, 3, 5-tris(bromomethyl) benzene, 'Abz' represents 4-aminobenzoic acid, and 'J' stands for D-Leucine. The selection of the linker will be explained below. In addition to these bicycles, we selected K(BCN)-linker-knottin-RGD (3a), cyclo-[K(K(BCN)-linker-)RGDf] (3b) and K(BCN)-linker-GRGDS (3c) as benchmark peptides.
For the synthesis of peptide-cyclooctyne conjugates, we chose an approach in which the integrin-binding sequence was coupled to a peptide linker in solid phase peptide synthesis. Pallarola et al explored the role of the linker system attached to cyclic RGD peptides in the inhibition of binding of immobilized vitronectin to the soluble integrin αvβ3 [50]; in their study, linkers comprising a triple- (HexPPP), hexa- (HexPPPPPP) or nonaproline (HexPPPPPPPPP) motif did not significantly decrease the inhibition ability. Hence, we added a linker that is similar to the HexPPP linker, but with some variations, to all the RGD peptides. In order to improve solubility in water-based solvents, we included two additional serine residues, and for practical reasons, we used 4-aminobenzoic acid instead of a triazole group. Furthermore, we incorporated an N-terminal acetylated lysine comprising a free amine in the side chain that allows for conjugation with BCN. Linear peptides comprising RGD in one loop and a tripeptide motif providing integrin-selectivity and additional affinity in the other, both enclosed by cysteine residues, were transformed into bicyclic peptides via trivalent scaffold 1, 3, 5-tris(bromomethyl)benzene followed by reaction with BCN-NHS to form bicyclic RGD peptide-cyclooctyne conjugates. A table of calculated and theoretical molecular weights obtained via UPLC/MS analysis can be found in the supporting information (table S1).
ELR functionalization and MALDI-TOF MS analysis
We subsequently functionalized the ELR azides with the peptide-cyclooctyne conjugates via copper-free click chemistry. This orthogonal reaction does not require catalysts and can be performed at room temperature in short reaction time (below one hour). Due to the need to maintain the ELRs in solution (below the Tt), the functionalization of ELRs and ELR azides was performed at 5 °C overnight. Six bicyclic RGD-peptides (1a–c, 2a–c) and three non-selective RGD-peptides (3a–c) were used to functionalize either 5% or 10% of the ELR azide groups to receive a total of 18 ELR-peptide conjugates (table 1). MALDI-TOF-MS analysis was performed to verify functionalization of the ELRs with the RGD peptides. Selected spectra (P0, P1a, P2a, and P3a–c) are shown in figure 2. The peaks at around m/z 62 200 represent non-functionalized ELR, whereas additional peaks reveal ELRs that were mono-, di-, tri- or even tetra-functionalized with RGD peptides (table 2).
Figure 2. MALDI-TOF MS spectra for non-functionalized ELR and ELRs functionalized with 5%/10% of bicyclic RGD peptides (1a), (2a) and control RGD peptides (3a–c).
Download figure:
Standard image High-resolution imageTable 2. Comparison between the different rates of functionalization and the peaks detected during MALDI-TOF MS analysis.
| 5% | Mono-functionalization |
| 10% | Di-functionalization |
| 15% | Tri-functionalization |
| 20% | Tetra-functionalization |
All the spectra in figure 2 reveal differences between 5% functionalization and 10% functionalization. In the spectra representing ELRs with 5% peptide functionalization, the peaks for monovalent functionalization have a higher intensity than those for bivalent functionalization. In contrast, the spectra representing ELRs with 10% peptide functionalization show different peak ratios, with the mono- and di-functionalization peaks showing higher intensity than the respective peaks for 5% functionalization. Moreover, additional peaks for tri- and tetravalent-functionalization of ELRs appear in the spectra for ELRs with 10% peptide functionalization.
Analysis of turbidity by UV/vis-spectroscopy
A turbidity study was performed for all the functionalized ELRs. In all the cases, a slight shift in Tt to lower values was observed as compared to the non-functionalized ELR (VKV-N3). The turbidity measurements are reported in the supporting information (figures S9–S11).
Contact angle
Static aqueous contact angle analysis was performed in order to verify the correct adsorption of ELRs and ELR-Peptides onto tissue culture polystyrene surfaces (TCPS). Measurements were taken using ultrapure water drops on four different surfaces: TCPS, VKV adsorbed on TCPS (VKV), VKV-N3 adsorbed on TCPS (VKV-N3), ELR bearing peptides adsorbed on TCPS (ELR-Peptide). The results (figure 3) show that TCPS is moderately hydrophobic (Ɵ = 71.6° ±0.5°), whereas VKV and VKV-N3 are markedly hydrophilic (Ɵ = 24.2° ± 1.5° and Ɵ = 23.3° ± 3.3°, respectively). ELR-Peptides was the most hydrophilic (Ɵ = 15.6° ± 1.6°), with the difference with respect to VKV-N3 being significant (p < 0.0001), thereby suggesting a hydrophilic contribution from the peptides. Measurements were repeated in different regions in order to confirm the homogeneous adsorption of ELRs and ELR-peptide onto TCPS.
Figure 3. Contact angle analysis performed 5 s after application of the drops to the surfaces: TCPS; VKV adsorbed on TCPS (VKV); VKV-N3 adsorbed on TCPS (VKV-N3); ELR-bearing peptides adsorbed on TCPS (ELR-Peptide). (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Download figure:
Standard image High-resolution imageXPS
XPS was used to characterize the TCPS surface adsorbed with ELRs and ELR-Peptides. The surface compositional results from XPS analysis, shown in table 3, indicate a reduction in C1s upon comparing untreated TCPS with VKV-N3 adsorbed on TCPS (VKV-N3) and ELR-bearing peptides adsorbed on TCPS (ELR-Peptide). XPS analysis also revealed a clear enrichment of nitrogen, passing from 0.8% for TCPS to 16.49% and 13.97%, respectively, for VKV-N3 and ELR-Peptide. Finally, oxygen also exhibited a slight increase compared with the TCPS surface.
Table 3. XPS analysis performed on different surfaces: TCPS; VKV adsorbed on TCPS (VKV); VKV-N3 adsorbed on TCPS (VKV-N3); ELR-bearing peptides adsorbed on TCPS (ELR-Peptide). All measurements were performed in triplicate. Values are expressed in % (average ± st. dev).
| C1s | N1s | O1s | |
|---|---|---|---|
| TCPS | 85.64 ± 1.82 | 0.74 ± 0.09 | 13.63 ± 1.73 |
| VKV | 68.69 ± 0.57 | 15.13 ± 0.28 | 16.18 ± 0.28 |
| VKV-N3 | 68.28 ± 1.29 | 15.71 ± 1.10 | 16.01 ± 0.20 |
| ELR-Peptide | 69.79 ± 0.20 | 14.33 ± 0.51 | 15.89 ± 0.32 |
Cell-adhesion assay
Time-dependent cell quantification studies
The capacity to promote cell adhesion and proliferation of HUVECs on the modified ELRs was evaluated at short, medium and long term. Figure 4 shows the time-dependent cell quantification for VKV-N3 (P0) and ELRs functionalized with 5% (P1a) and 10% modification (P1a-10). The cell count for all time points (except after 14 d) show a significant difference upon comparing (P0) with (P1a) and (P1a-10). However, no significant difference was observed between (P1a) and (P1a-10) at any time point. Similar behavior was found for the remaining ELR-peptides (supporting information, figure S7), and the various high-affinity integrin-binding peptides used in this study did not show any difference between the two concentrations used. As such, we decided to analyze the results for ELR-peptides functionalized at 5% (figures 5–7).
Figure 4. Time-dependent cell quantification for VKV-N3 (P0), P1a 5% (P1a) and P1a 10% (P1a-10) at different time points (30 min, 4 h, 1 d, 3 d, 5 d, 7 d, 14 d). All experiments were carried out in triplicate and error bars show standard deviations (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). There was no statistically significant difference between P1a and P1a-10 at any time point.
Download figure:
Standard image High-resolution imageFigure 5. Short-term time-dependent cell quantification (30 min, 4 h, 1 d). Number of cells determined for ELRs containing bicyclic peptides (1a–c) (red bars) and (2a–c) (green bars), and control RGD peptides (3a–c) (blue bars), RGD in the ELR-backbone (P0-RGD), ELR with no RGD peptide (P0), fibronectin coating (P0-FN) and coating with BSA (P0-BSA). The number of cells was calculated from fluorescence intensities using a calibration curve. All experiments were carried out in triplicate and error bars show standard deviations. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Download figure:
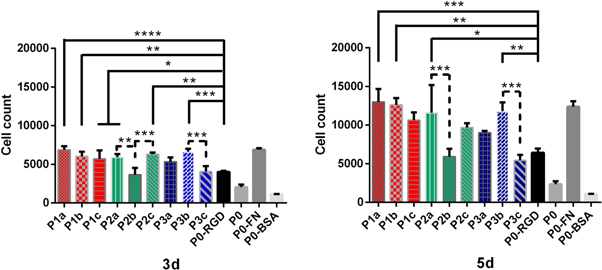
Standard image High-resolution imageFigure 6. Medium-term time-dependent cell quantification (3 d, 5 d). Number of cells determined for ELRs containing bicyclic peptides (1a–c) (red bars) and (2a–c) (green bars), and control RGD peptides (3a–c) (blue bars), RGD in the ELR-backbone (P0-RGD), ELR with no RGD peptide (P0), fibronectin coating (P0-FN) and coating with BSA (P0-BSA). The number of cells was calculated from fluorescence intensities using a calibration curve. All experiments were carried out in triplicate and error bars show standard deviations. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Download figure:
Standard image High-resolution imageFigure 7. Long-term time-dependent cell quantification (7 d, 14 d). Number of cells determined for ELRs containing bicyclic peptides (1a–c) (red bars) and (2a–c) (green bars), and control RGD peptides (3a–c) (blue bars), RGD in the ELR-backbone (P0-RGD), ELR with no RGD peptide (P0), fibronectin coating (P0-FN) and coating with BSA (P0-BSA). The number of cells was calculated from fluorescence intensities using a calibration curve. All experiments were carried out in triplicate and error bars show standard deviations. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Download figure:
Standard image High-resolution imageAll the PicoGreen® data collected were subdivided into different groups based on the different time points: short-term (30 min, 4 h, 1 d; figure 5), medium-term (3, 5 d; figure 6) and long-term (7, 14 d; figure 7). In order to make the bar charts clearer, the ELR-Peptides have been arranged and clustered into different groups: ELRs functionalized with αvβ3-binding bicycle peptides (1a–c) (red bars); ELRs functionalized with αvβ3 and α5β1-binding bicyclic peptides (2a–c) (green bars); and benchmark RGD peptides (3a–c) (blue bars). The grey scale bars represent other controls, namely the RGD in the ELR-backbone (P0-RGD), ELR with no RGD peptide (P0), fibronectin coating (P0-FN) and coating with BSA (P0-BSA). The positive (P0-FN) and negative controls (P0-BSA) remained between the highest and lowest, respectively, thus confirming the reliability of the study.
For the short-term study (30 min, 4 h, 1 d; figure 5), the peptides included after click modification supported higher levels of cells than the recombinant ELR-RGD (P0-RGD). At 30 min all the ELR-Peptides, except for P3c, showed the maximum statistical difference with (P0-RGD). The differences in cell numbers remained high for almost all ELR-Peptides when compared with the (P0-RGD) for the whole short-term study. In contrast, no significant difference was observed between the recombinant ELR-RGD (P0-RGD) and the ELRs lacking RGD (P0). The second comparison (indicated by a dashed line) shows the differences between the ELR-Peptide groups, in other words, bicyclic peptides (1a–c) (red bars), (2a–c) (green bars), and control RGD peptides (3a–c) (blue bars). Essentially no significant differences were found between the bicyclic peptide groups, with the only exception being for the group of non-selective RGD-peptides (3a–c), for which a greater variability and significance were evident. The ELR functionalized with (3a) supported cell adhesion most efficiently within this group, with cell counts similar to those for ELRs functionalized with integrin-selective bicycles (1a–c and 2a–c), whereas (3c) showed the lowest number of cells among the covalently functionalized ELRs at this time point, albeit still higher than for (P0-RGD).
The data collected at 4 h are similar to those obtained at 30 min, with all ELR-Peptides except (P3c) showing the same statistical difference with respect to (P0-RGD). The inter-group comparisons were in accordance with the trend observed at 30 min, with the bicyclic peptide groups (1a–c) (red bars), (2a–c) (green bars) showing no significant differences except for the group of non-selective RGD-Peptides (3a–c).
After 1 d, the differences between groups tended to decrease further, although the previous trends were maintained. A slight increase in cell counts at this time point suggests an incipient proliferation, although this is not particularly pronounced at this cell stage.
In the medium-term study (3 d, 5 d; figure 6), the number of cells practically doubled for almost all conditions, with a similar trend as for the initial adhesion, although with some exceptions. However, the comparison between the ELR-Peptides and (P0-RGD) appears less clear than for the short-term culture. Thus, despite showing good initial adhesion, (P2b) proliferation on slowed down over longer time periods. In contrast, the difference between (P0-RGD) and (P0) became more evident. A comparison within the same group revealed statistically significant differences only for (P2a–c) and (P3b–P3c) at day 3, and (P2a–P2b) and (P3b–P3c) at day 5.
Long-term proliferation studies (7 d, 14 d) revealed that HUVEC growth tends to reduce the difference in the ability to promote cell adhesion by the various high-affinity integrin-binding peptides (figure 7). Thus, after 7 d almost all the bicyclic peptides, i.e. (P1a–c), (P2a) and (P2c), showed a significant difference compared with (P0-RGD), whereas (P3a–c) and (P2b) exhibited no clear difference. No significant difference was found within ELR-Peptide groups. Finally, after 14 d all conditions except the negative control (P0-BSA) exhibited similar amounts of cells. Indeed, cell counts after 14 d were approximately threefold higher that after culturing for 7 d under all conditions.
Morphology studies
The ELR-Peptides were also characterized by morphological in vitro studies to investigate cell attachment, cell spreading, cytoskeletal reorganization and formation of focal adhesions. In order to investigate the time-dependent morphology of the cells, they were stained with rhodamin/phalloidin (actin skeleton), DAPI (nuclei) and a fluorescent mAb (vinculin) and examined by fluorescence microscopy (figures 8 and 9).
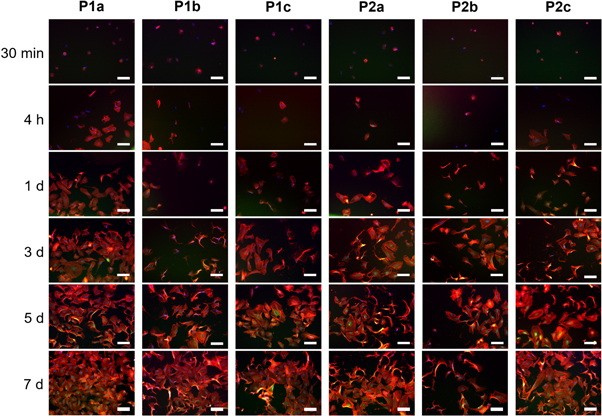
Figure 8. Immunostaining of actin (red), vinculin (green), and nuclei with DAPI (blue) of HUVEC cells cultured for different time points (30 min, 4 h, 1 d, 3 d, 5 d, 7 d) on ELRs functionalized with bicycles (1a–c) (P1a–c) and (2a–c) (P2a–c). Scale bars: 100 μm.
Download figure:
Standard image High-resolution imageFigure 9. Immunostaining of actin (red), vinculin (green), and nuclei with DAPI (blue) of HUVEC cells cultured for different time points (30 min, 4 h, 1 d, 3 d, 5 d, 7 d) on ELRs functionalized with controls (3a–c) (P3a–c), and comprising RGD in the ELR-backbone (P0-RGD), no RGD peptide (P0), fibronectin coating (P0-FN) and coating with BSA (P0-BSA). Scale bars: 100 μm.
Download figure:
Standard image High-resolution imageSince non-adhered cells were discarded after seeding for 30 min, those that remained adhered, although small and spherically shaped, nevertheless showed small protrusions at the periphery or ring-shaped adhesions at the onset of cell culture (30 min) under practically all conditions tested (supporting information, figure S8). At early time points, the focal contacts are circumscribed to the perinuclear zone, whereas at longer times a yellow coloration was found when overlapping actin and vinculin captures.
The quantitative image analysis of cell spreading area has been performed after short time incubation (4 h) (figure 10). (P1a–b), (P2c) and (P3a) showed the highest statistical difference as compared with (P0-RGD), whereas the difference between (P3b) and (P2a) was less significant. All the remaining ELR-Peptides (P1c, P2b and P3c) did not exhibit significantly different cell spreading as compared with (P0-RGD). In any case, no significant differences were found within the same peptide group. (P2b) showed the largest error bars, reflecting the vast population of cells having a different spreading area (figure 10). In contrast, the negative control (P0-BSA) does not appear since no cells were present, (figure 9). The data collected by quantitative image analysis after 4 h are similar to those obtained by cell quantification analysis reported in figure 5. In both analyzes, (P2b) and (P3c) showed less difference as compared with (P0-RGD). With respect to the cell spreading area analysis, also P3b and P2a showed less significant difference compared to (P0-RGD), whereas for (P1c) no significant differences were found. When comparing cell spreading areas on ELRs within the same group, the trends observed were in accordance with cell quantification analysis, where no significant differences were shown, except for the group of non-selective RGD-Peptides (3a–c).
Figure 10. Quantitative image analysis of cell spreading area after 4 h. Cell area has been determined for ELRs containing bicyclic peptides (1a–c) (red bars) and (2a–c) (green bars), and control RGD peptides (3a–c) (blue bars), RGD in the ELR-backbone (P0-RGD), ELR with no RGD peptide (P0) and fibronectin coating (P0-FN). A minimum of 25 images have been analyzed per sample, except for (P1c), (P2b), (P3b–c) and (P0) where the number of images was less than 25 but in any case more than 10. The results correspond to a single independent experiment. The values are is expressed μm2; the error bars show standard deviations. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Download figure:
Standard image High-resolution imageDiscussion
The focus of biomaterials research often lies with the biomaterial itself rather than the cell-adhesion sequence [51], especially given that the surface-protein interaction determines the nature of subsequent cell-surface behavior. Herein we have created peptide-functionalized ELRs by conjugating various high-affinity integrin-binding, bicyclic RGD-Peptides to ELRs via copper-free click chemistry in order to overcome the known limitations of recombinant synthesis, for instance, the inability to incorporate non-canonical amino acids and to form cyclized peptides. Furthermore, this strategy allows the bioactive ligand to maintain the flexibility and minimal steric hindrance required for cellular interactions. The purpose of this study was to improve the cell-adhesion and proliferation abilities of the recombinant and biocompatible substrate ELRs by conjugating a very small number (one or two molecules per ELR) of high integrin affinity peptides. The choice of the different conditions and peptides was based on very recent studies by Bernhagen et al who reported an exhaustive investigation of high-affinity integrin αvβ3- and α5β1-binding bicyclic RGD-Peptides [42, 43]. Remarkably, in the same studies the researchers also found that linear GRGDS (non-functionalized version of (3c)), which is probably the most common cell-adhesive ligand in hydrogels, showed relatively low integrin αvβ3 and α5β1 affinity, whereas the monocyclic peptide cyclo-KRGDf (non-functionalized equivalent to (3b)) showed a high affinity for integrins αvβ3 and α5β1. Similarly, the 32-mer knottin-RGD peptide (origin of (3a)) published by Kimura et al [52] non-selectively bound all integrins αvβ3, αvβ5 and α5β1 with high affinity. Finally, a battery of nine RGD-Peptides was successfully synthesized and conjugated with cyclooctyne using an approach in which the integrin-binding sequence was coupled to a peptide linker. Moreover, and discussed above, the solubility in water-based solvents was improved by the inclusion of additional serine residues. All the cyclooctyne-functionalized peptides were conjugated to ELR azides via copper-free click chemistry. Functionalization was carried out as a mono-functionalization (5%) and di-functionalization (10%), and the ELR-Peptides were further analyzed by MALDI-TOF MS. Although MALDI-TOF MS analysis does not allow the degree of functionalization to be determined quantitatively, a comparison of the spectra in figure 2 reveals different degrees of functionalization depending on the type and amount of peptide applied, thus confirming the successful conjugation of two different quantities of peptides on the ELRs. Furthermore, a turbidity study was performed for all the functionalized ELRs, with a slight shift in Tt to lower values beings observed for all ELR-Peptides in comparison with the non-functionalized ELR. This essentially negligible Tt shift is likely due to the very small quantity of peptide molecules conjugated to the ELRs (one molecule for 5% functionalization and two molecules for 10% functionalization) and to the low molecular weight of the peptides. These data show how this functionalization strategy does not affect the physical properties of the ELR, represented by the Tt value (supporting information, figures S9–S11).
The ELRs comprising 5% or 10% peptide functionalization were adsorbed onto TCPS in order to further investigate the cell adhesion behavior and proliferation over these surfaces. XPS and CA analysis were used to confirm the correct adsorption. Thus, XPS analysis revealed a change in the surface composition, as can be seen from table 3 for ELRs and ELR-Peptides, with a clear enrichment in nitrogen and a reduction in carbon, and oxygen increasing slightly compared with the TCPS surface. These changes in chemical composition confirm an adequate adsorption of ELRs and ELR-Peptides onto TCPS [53–55]. CA measurements help to characterize the affinity of a solution or suspension towards a certain surface, with the CA value decreasing as the number and strength of these interactions increases. The adsorption of ELRs onto the surface increases the hydrophilicity with respect to TCPS and, given the XPS results, this is likely due to the enrichment in electronegative atoms like nitrogen and oxygen and to the reduction in carbon. This enrichment in electronegative atoms increases the number and strength of interactions, such as hydrogen bonds, with the aqueous solution. Surprisingly, the ELR containing peptides adsorbed on TCPS was the most hydrophilic surface, with the difference with respect to the other ELRs being significant. This suggests a hydrophilic contribution from the peptides, which likely expose their electronegative atoms outwards, thereby increasing the number and strength of interactions with the aqueous solution. As reported previously, the hydrophilicity of a solid surface is an essential requirement for cell-surface interactions [56] and, in this case, combines with a correct exposure of RGD peptide sequences.
HUVECs were chosen to investigate the cell adhesion behavior, proliferation and morphology by culture thereof on ELR and ELR-Peptide adsorbed surfaces. Endothelial cells form part of a highly specialized tissue for vessel formation that provides stable structural support for new vessels. Endothelial cells cover the blood vessels and are metabolically very active, being responsible for maintaining vascular homeostasis. In this study, we have synthesized a battery of nine peptides which, can be sub-divided into groups of three with high affinity for αvβ3, α5β1/αvβ3, and multiple integrins (table 1). According to the literature, the remodeling of blood vessels and concomitant reorganization of the cytoskeleton requires the involvement of integrins [57]. In mammals, this family of integrins comprises at least 20 different αβ heterodimers, which are expressed on the surface of endothelial cells [57]. Different combinations of integrin subunits on the cell surface allow cells to recognize and respond to a variety of extracellular matrix proteins under different physiological conditions; for example, the αvβ1 and α5β1 fibronectin receptors are highly expressed in quiescent endothelial cells, whereas the αvβ3 fibronectin and vitronectin receptor is expressed only during angiogenesis [57, 58]. Herein we have evaluated either the functional avidity or proliferation of HUVECs on ELR and ELR-peptide adsorbed surfaces. Loosely adhered or unbound cells were removed from the surfaces after incubation for 30 min in order to determine the cell fate as a function of the extent of initial attachment to the different surfaces and, therefore, the avidity of the cells for the adhesion sequences exposed.
The adhesion behavior was investigated in a short-term study (up to 1 d, figure 5), which revealed that ELR-bearing peptides supported higher levels of cells than the recombinant ELR-RGD. This difference involves either the bicyclic peptides (1a–c) or (2a–c), each of which exhibits high-affinity for αvβ3 and α5β1/αvβ3 integrins, or the control RGD peptides, which exhibit high affinity for multiple integrins (3a–c). It should be noted that the backbone-RGD functionalized ELR (P0-RGD) comprises six RGD motifs per molecule, while the covalently functionalized ELR-Peptides only comprise one RGD moiety per molecule. This suggests either a higher adhesion capacity for the high-affinity integrin-binding peptides to HUVECs or a better exposure of these peptides after protein adsorption. It is well known from the literature that cell attachment is influenced by several factors, one of the most important of which is the RGD concentration [59–61]. By creating RGD-Peptides functionalized via copper-free click chemistry, we have improved the cell attachment capacity while reducing the RGD concentration on the surface, although the RGD-concentration function varies for each different bioactive surface and cell line [62, 63].
Surprisingly, the adhesion and proliferation results for the different high-affinity integrin-binding peptides used in this study showed no difference when compared with the same ELR-Peptide for the two concentrations selected. This identical behavior for ELRs having 5% and 10% peptide functionalization is likely due to the similar and very low number of peptide molecules (one and two molecules for 5% and 10%, respectively) on the ELR substrate. In contrast, the presence of RGD in the backbone (P0-RGD) did not affect the adhesion behavior of cells in the short-term culture when compared with the ELR lacking RGD (P0). This could be related to the adsorption process of ELR-RGD molecules, which may have not allowed an optimal outwards exposure of the RGD sequence. Overall, the short-term results demonstrate a greater avidity of the cells for the bicycle-functionalized ELRs. According to the cell adhesion and proliferation results obtained for ELR-Peptides functionalized at 5%, the differences observed in the short-term cultures could be explained by considering that signal transduction after interaction with a specific integrin can trigger different or preferential cellular responses (proliferation, migration and/or organization into networks characteristic of early angiogenesis, for example). A comparison within the same groups of ELR-Peptides also revealed clear differences for various peptides at certain time points. However, these differences do not follow a trend as they arise due to the design and synthesis of the different peptides which, although they have the same composition, exhibit different affinities for the integrins expressed on the HUVEC membrane. The PicoGreen® study is in accordance with the morphological study, whereby the focal contacts, which are initially circumscribed to the perinuclear zone, give way to vinculin clustering of activated integrins, as can be inferred from the yellow coloration found when overlapping actin and vinculin captures. In addition, these were numerous and not restricted to the nuclear periphery. The morphological study reveals that short-term capability to support cell adhesion is higher for the surface adsorbed with bicycle-functionalized ELRs than for all the other benchmarks. Moreover, the cell spreading area at a short time incubation (4 h) followed the same trend of cell adhesion, suggesting that bicyclic RGD peptides support cell adhesion and cell cytoskeleton.
The differences in cell numbers were highest in the short time study for almost all the peptide-bearing ELRs when compared with the recombinant ELR-RGD. However, a comparison between the ELR-Peptides and ELR-RGD showed much smaller differences in the long term, with HUVEC growth tending to minimize the initial difference in the ability to promote cell adhesion by the various high-affinity integrin-binding peptides. Differences within the same groups of ELR-Peptides were also relatively small. Intercellular contacts between HUVECs inhibit their proliferation and growth stops. A possible explanation for the similar cell numbers in long-term cultures (7 d, 14 d, figure 7) is that the rapid proliferation corresponds to an earlier confluence with respect to the cells that do not exhibit intercellular contacts. However, in contrast to the group of non-selective RGD-Peptides, which exhibit high affinity for multiple integrins, almost all the bicyclic peptides exhibited the highest proliferation data at longer times, showing a significant difference with respect to ELR-RGD. The cell quantification data show that essentially similar proliferation rates were observed for the bicyclic peptides optimized for both αvβ3 and α5β1 affinity/selectivity, thus enabling either active or quiescent HUVECs to be targeted. The morphological study shows how the surfaces that have performed a better adhesion at short times, are those where the cells exhibit more expanded and less sharped morphology at longer times. Given that a similar situation is found for cells cultured on fibronectin (positive control), the peptide-bearing ELRs appear to be a better substrate for cell stretching compared with the recombinant-synthesized ELR-RGD (P0-RGD) and ELR itself (P0). However, these differences became less apparent as cell cultures were maintained for longer periods, probably as a result of the analysis chosen for this cell type.
In light of these results, the various high-affinity integrin-binding peptides used in this study appear to play an important role, especially during short-term culture. RGD Peptide-functionalized ELRs enhance the ability to promote cell adhesion compared with recombinant-synthesized ELR-RGD, irrespective of the lower net RGD concentration of covalently functionalized ELRs. Moreover, taking into account the limitations of in vitro culture, an in vivo study would be required to determine whether active HUVECs involved in angiogenesis could be selectively targeted with bicycle-functionalized ELRs optimized for high αvβ3 integrin affinity (P1a–P1c). Similarly, bicycle-functionalized ELRs optimized for α5β1 affinity (P2a–P2c) could exhibit a specific interaction with endothelial cells in a quiescent state. Finally, we have opened up a new application for ELRs as a biocompatible substrate in studies to determine which integrins need to be targeted for optimal cell adhesion and proliferation.
Conclusion
The in vitro studies of ELRs functionalized with high-affinity integrin αvβ3- and α5β1-binding RGD bicycles suggest that these bicycles provide an interesting alternative to promote fast cell adhesion on 2D biomaterial surfaces compared with well-known linear or monocyclic RGD peptides. Our initial hypothesis that high-affinity integrin-binding RGD bicycles, as determined in solid-phase immunoassays [43], should improve integrin-mediated cell adhesion and proliferation to a significantly greater extent than monocyclic and linear RGD was partially verified. Furthermore, we have shown that covalent RGD-functionalization of ELRs via copper-free click reaction is more efficient for inducing integrin-mediated cell adhesion and proliferation than the recombinant synthesis of ELRs comprising RGD as part of their backbone. This strategy could be used to ensure correct exposure of the bioactive sequence, thereby guaranteeing an optimal cell-material interaction. Finally, we believe that ELRs functionalized with integrin-selective RGD-bicycles represent an attractive and efficient way to design integrin-selective polymer surfaces with the potential to evaluate cell-adhesion behavior and tailor high integrin peptides for specific biomedical applications.
Acknowledgments
This project has received funding from the European Union'sHorizon 2020 research and innovation programme under theMarie Sklodowska-Curie grant agreement No. 642687. The authors are grateful for the funding from the European Commission (NMP-2014-646075), MINECO of the Spanish Government (PCIN-2015-010, MAT2015-68901-R, MAT2016-78903-R, MAT2016-79435-R), Junta de Castilla y León (VA317P18) and Centro en Red de Medicina Regenerativa y Terapia Celular de Castilla y León.
Notes
Pepscan is the inventor of the CLIPS technology and holds a patent on the synthesis of bicyclic peptides using this technique.