Abstract
The biological membrane is a dynamic supramolecular architecture that plays vital roles in the cell. However, understanding the physicochemical properties and functions of the membrane supramolecular system is difficult. We have developed an integrated model system of the biological membrane comprising patterned polymeric and natural lipid bilayers. The polymeric bilayer acts as a framework to support embedded natural membranes. The embedded natural membranes retain important characteristics of the biological membrane such as fluidity, and reproduces the physical states and functions of the biological membrane. Membrane proteins can be reconstituted into the model membrane for analyzing their functions in a controlled lipid membrane environment. Three-dimensional structures can be constructed by attaching micro-/nano-fabricated structures to the polymeric bilayer framework. The integrated model membrane realizes a versatile platform to study membrane functions, and should open new opportunities in fundamental biological sciences as well as biomedical/analytical applications.
Export citation and abstract BibTeX RIS
1. Introduction
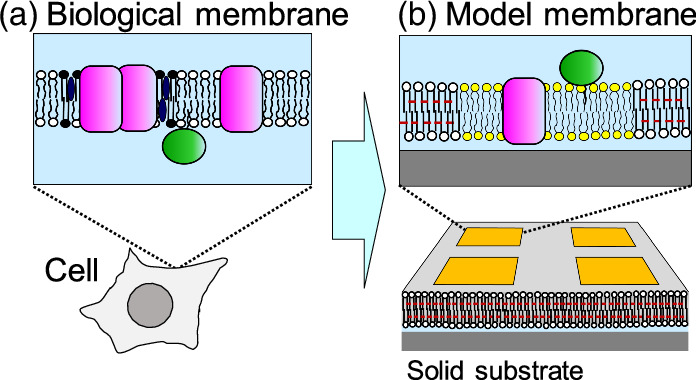
Biological membranes composed of lipids and proteins are ubiquitous in the cell. They are closely related with a wide range of cellular functions, spanning from signal transduction to energy harvest by photosynthesis. Diverse membrane functions derive from their dynamic and complex supramolecular structures and unique physicochemical properties. Biological membranes are a two dimensional (2D) fluid, in which lipids and proteins can diffuse laterally. The molecules in the membrane are not homogeneously distributed but form heterogeneous domains (e.g. lipid raft, protein clusters) [Fig. 1(a)]. 1) The 2D distribution of molecules should play important roles in the membrane functions. The membrane also defines the three dimensional (3D) border of cell interior and exterior, and further divide the cell into organelles. Formation of nanoscopic compartments is one of the key features of the membrane. Studying the structures, physicochemical properties, and functions of the biological membrane is challenging owing to its dynamic and complex supramolecular architecture. As an approach to understand the machinery of biological membranes, in-vitro assay systems composed of defined set of molecules have been developed, including lipid vesicle (liposome), black lipid membrane, and supported lipid bilayer (SLB). These bottom-up approaches have provided insight into the structural and functional characteristics of the biological membrane, laying the foundation of our current understanding. 2)
Fig. 1. Scheme of the biological membrane and patterned model membrane. (a) Lipid molecules have hydrophilic and hydrophobic sections in a single molecule (represented with a circle and two tails, respectively), and form a bilayer structure. Membrane proteins either span the bilayer membrane (pink molecules) or adsorb on the surface of the membrane (green molecules). (b) A patterned model membrane reproduces the essential features of the biological membrane on a solid substrate.
Download figure:
Standard image High-resolution imageSince SLB is supported on a planar solid substrate such as silicon and glass, interfacial analytical methods and micro-fabrication techniques can be applied. 3,4) Utilizing these unique features of SLBs, we have developed an integrated model system of the biological membrane comprising patterned polymeric and natural lipid bilayers [Fig. 1(b)]. 5,6) The polymeric bilayer acts as a framework to support embedded natural membranes. The embedded natural membranes retain important characteristics of the biological membrane such as fluidity, and reproduces the functions of the biological membrane. The patterned model membrane can mimic the lateral organization of membrane-bound molecules and vertical structures of membrane nano-compartments. By mimicking key membrane features and functions, we envision that the integrated model membranes can be applied to basic membrane studies as well as biomedical applications.
2. Experimental methods
2.1. Lithographic patterning of lipid bilayer membrane
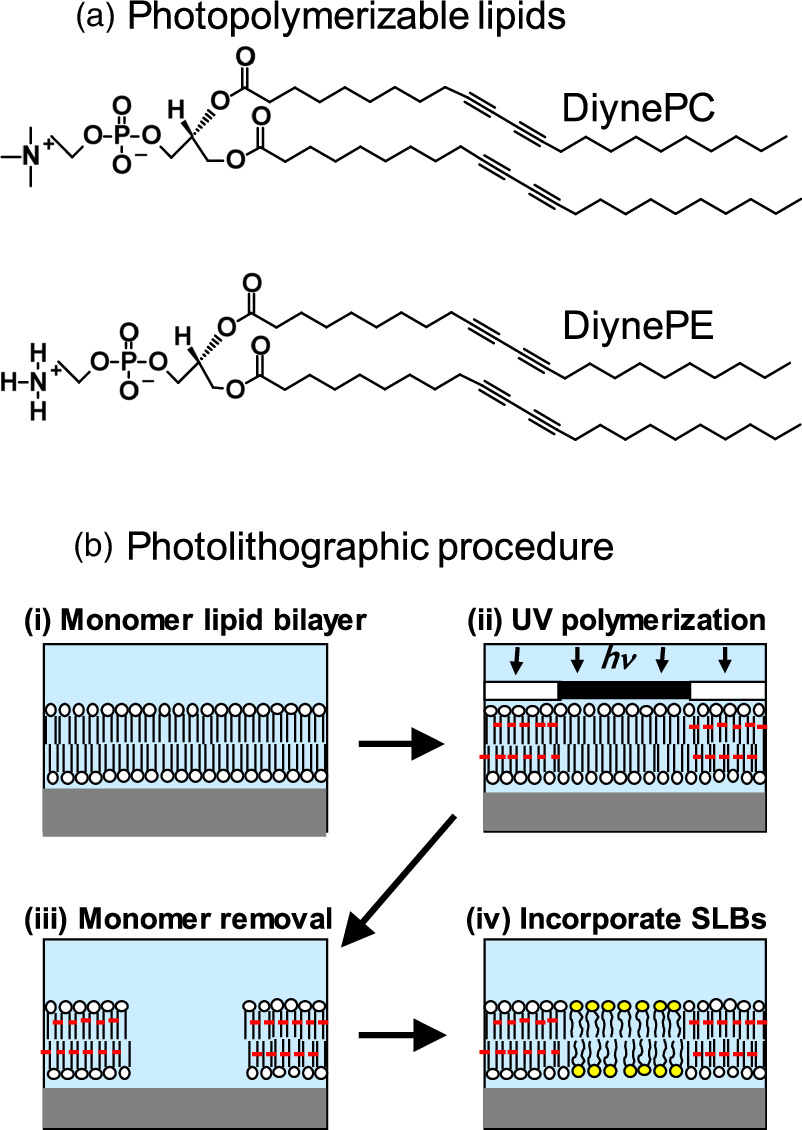
Patterned SLBs were generated on a microscopy coverslip by photolithographic polymerization of 1,2-bis(10,12-tricosadiynoyl)-sn-glycero-3-phos-phocholine (DiynePC) (Fig. 2). 7) Microscopy coverslips were cleaned with 0.1 M sodium dodecyl sulfate (SDS) for 20 min under sonication, rinsed with deionized water, treated in a solution of NH4OH (28%)/H2O2 (30%)/H2O (0.05:1:5) for 10 min at 65 °C, rinsed extensively with deionized water, and then dried under nitrogen blow. The substrates were further cleaned by the UV/ozone treatment for 20 min (PL16−110, Sen Lights Corporation, Toyonaka, Japan). Bilayers of monomeric DiynePC were deposited onto substrates by the spontaneous transformation of lipid vesicles into SLB (vesicle fusion). The substrate was cooled on ice to induce rapid formation of crystalline domains. 8) Polymerization of DiynePC bilayers was conducted by UV irradiation using a Mercury lamp (SP9, Ushio, Tokyo, Japan) as the light source. The applied UV intensity was typically 170 mW cm−2 at 254 nm, and the UV dose was modulated with the illumination time. After UV irradiation, nonpolymerized DiynePC molecules were removed from the substrate surface by immersing in 0.1 M SDS at 30 °C and rinsing extensively with deionized water. Natural lipid bilayers were incorporated into the patterned framework of polymeric bilayer by vesicle fusion or spontaneous spreading of phospholipids from lipid aggregates (self-spreading). 9,10)
Fig. 2. Fabrication of patterned lipid bilayers: (a) structures of photopolymerizable lipids: DiynePC and DiynePE have a polymerizable diacetylene moiety in the hydrocarbon chains. DiynePE has a chemically reactive amine moiety in the hydrophilic head group. (b) Photolithographic fabrication scheme: (i) deposition of monomeric SLB by vesicle fusion or LB/LS, (ii) UV polymerization with a photomask, (iii) removal of unreacted monomer with a detergent solution (0.1 M SDS), and (iv) incorporation of natural lipid membranes (lipids with yellow circles).
Download figure:
Standard image High-resolution imageMembrane proteins were reconstituted into the patterned membrane either from purified/solubilized micelles or crude native membranes. The photoreceptor rhodopsin (Rh) was purified from the rod outer segments (ROS) of bullfrogs (Rana catesbeiana), and was reconstituted into a pre-formed SLB by the rapid dilution of solubilized Rh molecules. 11) In a different set of experiment, native membranes such as thylakoids from spinach chloroplast were reconstituted into the preformed polymeric bilayer framework by applying them together with phospholipid vesicles. 12) The phospholipid vesicles acted as a catalyst that induced spontaneous reorganization of thylakoid membranes into a planar SLB surrounded by polymeric bilayers.
2.2. Biomimetic 3D architectures based on patterned lipid membrane
To realize biomimetic 3D architecture based on a patterned membrane, the surface of polymeric bilayer was functionalized using a polymerizable phospholipid bilayers having a reactive ethanolamine head group 1,2-bis(10,12-tricosadiynoyl)-sn-glycero-3-phosphoethanolamine (DiynePE) (Fig. 2, Fig. 3). Although DiynePE does not form a stable bilayer structure because the head group is significantly smaller than the hydrophobic tails, 13) stable bilayers could be formed by mixing DiynePE with a polymerizable phospholipid having a choline head group (DiynePC). Bilayers composed of DiynePC and DiynePE were deposited onto substrates from the air/water interface by the Langmuir–Blodgett (LB) and subsequent Langmuir–Schaefer (LS) methods using a Langmuir trough (HBM-AP, Kyowa Interface Science, Asaka, Japan). The LB/LS process was employed, since formation of SLBs by vesicle fusion is unfavorable for DiynePC/DiynePE due to the intrinsic curvature of PE-containing membrane. 14) Lithographic polymerization of DiynePC/DiynePE bilayers was conducted by UV irradiation and subsequent SDS treatment as described above. Natural lipid bilayers were incorporated into the patterned framework of polymeric bilayer by vesicle fusion.
Fig. 3. Extending the patterned membrane into the vertical direction (3D architecture): (A) functional molecules can be conjugated to the surface of polymeric bilayer using chemically reactive DiynePE. The lipid molecules are represented with a circle and two tails. Lipid molecules with yellow, white, and red circles are natural lipids, DiynePC, and DiynePE, respectively. (B) Various objects, including biomolecules (depicted as a triangle with a linking chain) (a), cells (b), and (c) micro-structures (e.g. PDMS micro-channels, chambers), can be attached to the functional molecules on the polymeric bilayer.
Download figure:
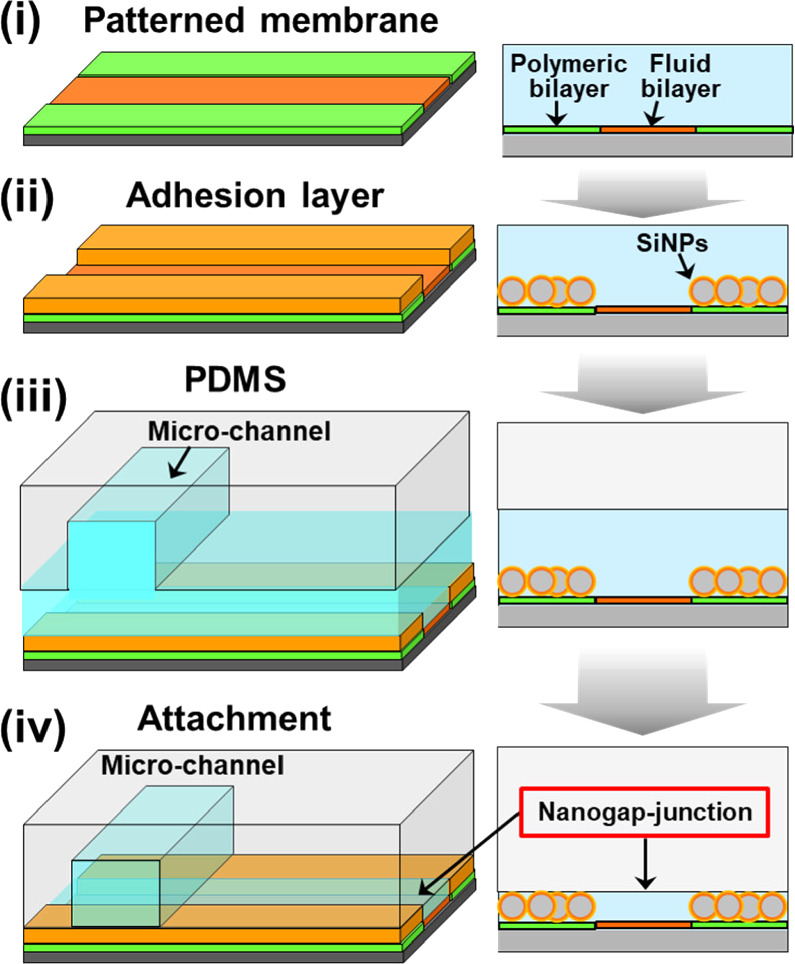
Standard image High-resolution imageThe surface of DiynePC/DiynePE bilayer was functionalized with biotin using a conjugate of N-Hydroxysuccinimide (NHS), polyethylene glycol (PEG), and biotin (NHS-PEG4-biotin). A nanometer-sized gap-junction was generated by attaching a patterned bilayer with a polydimethylsiloxane (PDMS) slab using an adhesion layer having a defined thickness (Fig. 4). As the adhesion material, we used polymer-coated silica nanoparticles (SiNPs). Polymer brush consisting of 2-methacryloyloxyethyl phosphorylcholine (MPC) and 2-aminoethyl methacrylate (AEMA) (poly(MPC-co-AEMA) was formed on SiNPs by the surface-initiated atom transfer radical polymerization (ATRP). 15) To use the polymer-coated SiNPs as the adhesion material, AEMA in the polymer brush was biotinylated. Patterned polymeric bilayer, SiNPs, and PDMS surfaces were bound with the biotin-streptavidin linkage.
Fig. 4. Fabrication of nanogap-junction between a patterned membrane and PDMS using silica nanoparticles as the adhesion layer. (i) Patterned bilayer, (ii) the adhesion layer is attached onto the surface of the polymeric bilayer, (iii)–(iv) PDMS is attached onto the adhesion layer, forming a gap structure between the fluid bilayer and PDMS. It should be noted that all processes were conducted in water (depicted with blue color). Figure 4 was modified from M. Tanabe et al., Small. 14, 1802804 (2018) with permission of Wiley.
Download figure:
Standard image High-resolution image3. Results and discussion
3.1. Patterned membrane composed of polymeric and fluid lipid bilayers
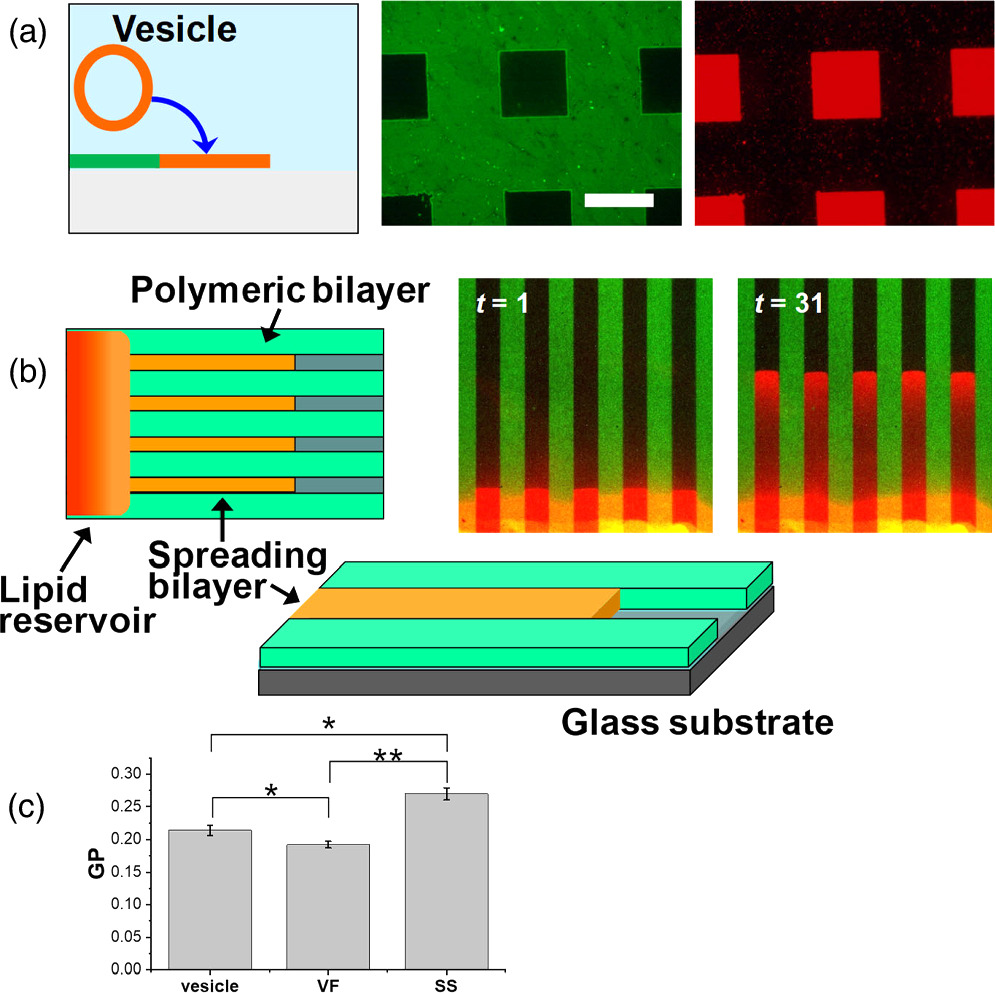
Lithographic polymerization of DiynePC bilayer and subsequent incorporation of natural lipid bilayers result in a patterned hybrid membrane (Fig. 2). The polymeric bilayer acts as a framework for defining the area of natural lipid bilayers, whereas the natural lipid bilayers retain the physicochemical properties of biological membrane such as lateral mobility of membrane-bound molecules (fluidity). Natural lipid bilayers were incorporated into the patterned polymeric bilayer scaffold by two spontaneous processes, i.e. vesicle fusion and self-spreading (Fig. 5). 9,10) In vesicle fusion, lipid vesicles adsorb onto a hydrophilic surface, rupture, and lipid patches fuse with each other. Currently, SLBs are most commonly generated by vesicle fusion owing to its technical simplicity. 16,17) On the other hand, self-spreading is a spontaneous wetting phenomenon of a hydrophilic surface by a lipid bilayer that extends from a reservoir of liquid crystalline lipid bilayers. 18–20) This technique is potentially useful for the formation of supported membranes that are more resistant towards defects formation, since lipid molecules are continually supplied from the lipid reservoir. Heightened lipid packing was observed in self-spread bilayers compared with those formed by vesicle fusion [Fig. 5(c)]. These features give self-spreading important advantages for preparing patterned model membranes for reconstituting membrane proteins, where detergent solutions are applied. Importantly, the scaffold of polymeric bilayer can stabilize incorporated fluid lipid bilayers. Formation of SLBs was accelerated in the presence of polymeric bilayers both for vesicle fusion and self-spreading, indicated that the hydrophobic edges of polymeric bilayer reduced the energetic barriers of SLB formation. 9,10)
Fig. 5. Incorporation of natural lipid bilayers by (a) vesicle fusion and (b) self-spreading. (c) Lipid packing in vesicles and SLBs formed by self-spreading (SS) and vesicle fusion (VF) was evaluated with a fluorescent dye Laurdan, whose fluorescence is sensitive to the polarity of environment. Fluorescence of Laurdan was collected at two wavelength, and the normalized difference between the two emission intensities (general polarization: GP) was calculated. Higher GP indicates a higher packing of lipid molecules. The statistical significance of the difference: * P < 0.05 (significant), ** P < 0.01 (highly significant). Figure 5 was modified from K. Morigaki et al., Langmuir. 20, 7729 (2004) and F. Tamura et al., Langmuir. 35, 14696 (2019) with permission of American Chemical Society.
Download figure:
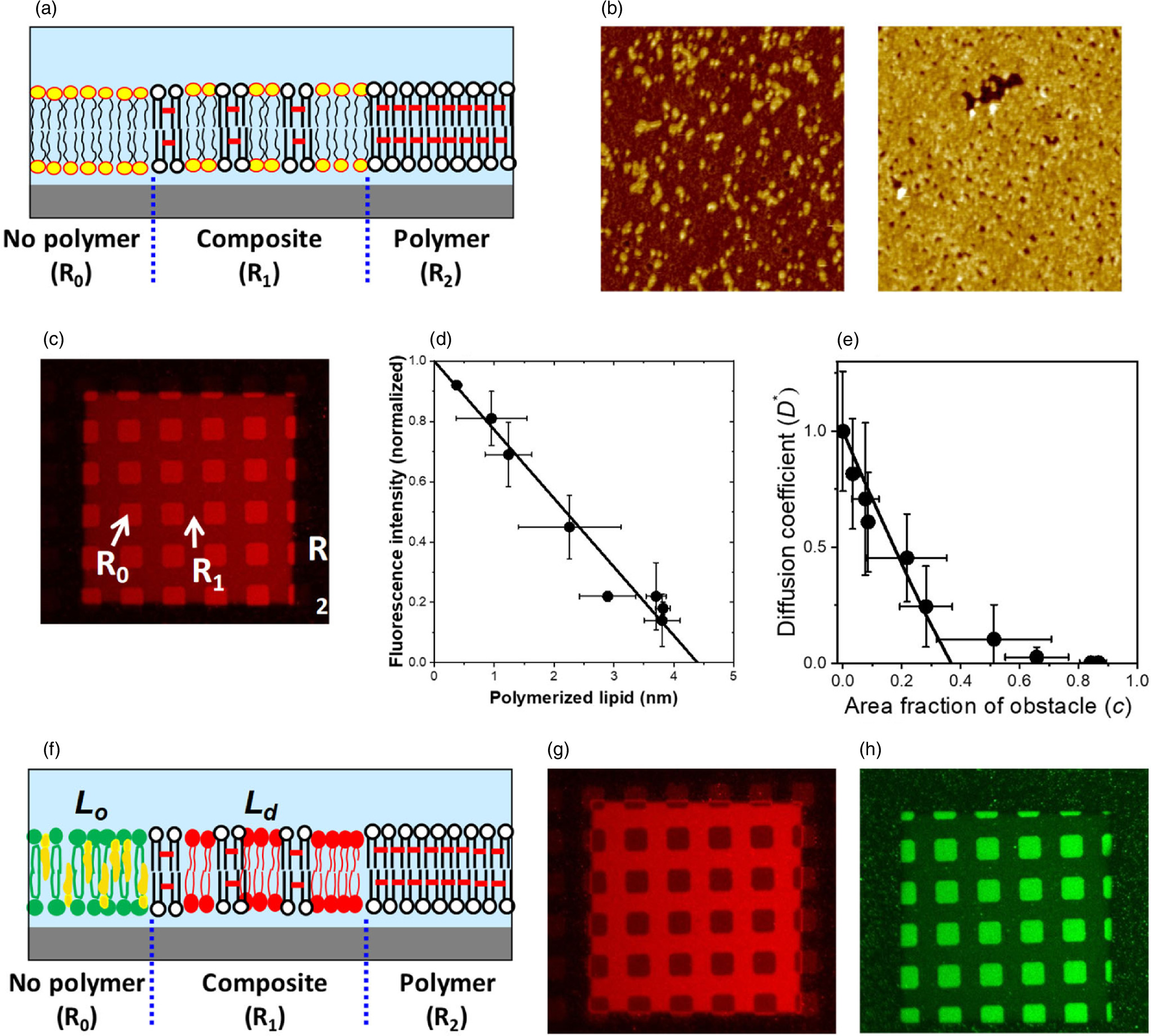
Standard image High-resolution imageIn the patterned hybrid membrane, polymeric and fluid lipid bilayers form a continuous membrane. A partially polymerized bilayer, in which nanoscopic polymeric bilayer domains only partially cover the substrate surface, can be generated by modulating the UV dose for polymerization [Figs. 6(a), 6(b)]. Fluid lipid bilayers can penetrate into the partially polymerized region, forming a nanometer-sized composite membrane of polymeric and fluid bilayers [Figs. 6(c), (d)]. 21,22) Lateral diffusion of membrane-bound molecules is obstructed by polymeric bilayer domains [Fig. 6(e)]. The degree of obstruction increases linearly with the polymeric bilayer coverage, and leads to percolation. An important manifestation of the partially polymerized bilayer region is spontaneous phase separation of lipid domains. As we introduced a lipid bilayer that can separate into liquid-ordered (Lo) and liquid-disordered (Ld) domains, we observed that Lo and Ld domains preferentially accumulated in the polymer-free and partially polymeric regions, respectively [Figs. 6(f)–6(h)]. 22,23) Two possible mechanisms are involved in the patterned phase separation. First, Ld domains preferentially accumulate at the boundary between polymeric and fluid bilayers due to the lower bending elasticity of Ld domains, which results in a reduced energetic cost of bilayer deformation. 24) The second factor is the Ostwald ripening of domains. Lo domains in the matrix of a continuous Ld phase tend to grow in size in order to minimize the line tension. 25) However, the domain growth is hindered in the partially polymerized regions by polymeric bilayer domains. Patterned Lo/Ld domains have cholesterol-enriched and cholesterol-depleted domains in a predefined geometry. They can be used as a model for lipid micro-/nano-domains in the biological membrane (e.g. lipid raft) for studying their physicochemical and functional roles.
Fig. 6. Formation of partially polymeric bilayer and patterned lipid domain separation. (a) Schematic of partially polymeric bilayer: R0, R1, R2 are the regions of non-polymer (natural), composite (polymer/natural), and polymeric bilayers, respectively. (b) AFM observation of polymer domains, (c) fluorescence of incorporated fluid bilayer (DOPC/TR-PE) in three regions, (d) fluorescence intensity versus average polymer thickness, (e) diffusion coefficient of TR-PE versus area fraction of obstacle (polymer). Patterned phase separation: (f) Schematic drawing, (g), (h) fluorescence micrographs of a phase-separated membrane observed with Rho-DOPE (Ld marker: G) and CTB-488 (Lo marker: H). Figure 6 were modified from T. Okazaki et al., Langmuir. 25, 345 (2009) and F. Okada et al., RSC Adv. 5, 1507 (2015) with permission of American Chemical Society and Royal Chemical Society, respectively.
Download figure:
Standard image High-resolution image3.2. Reconstitution and characterization of membrane proteins
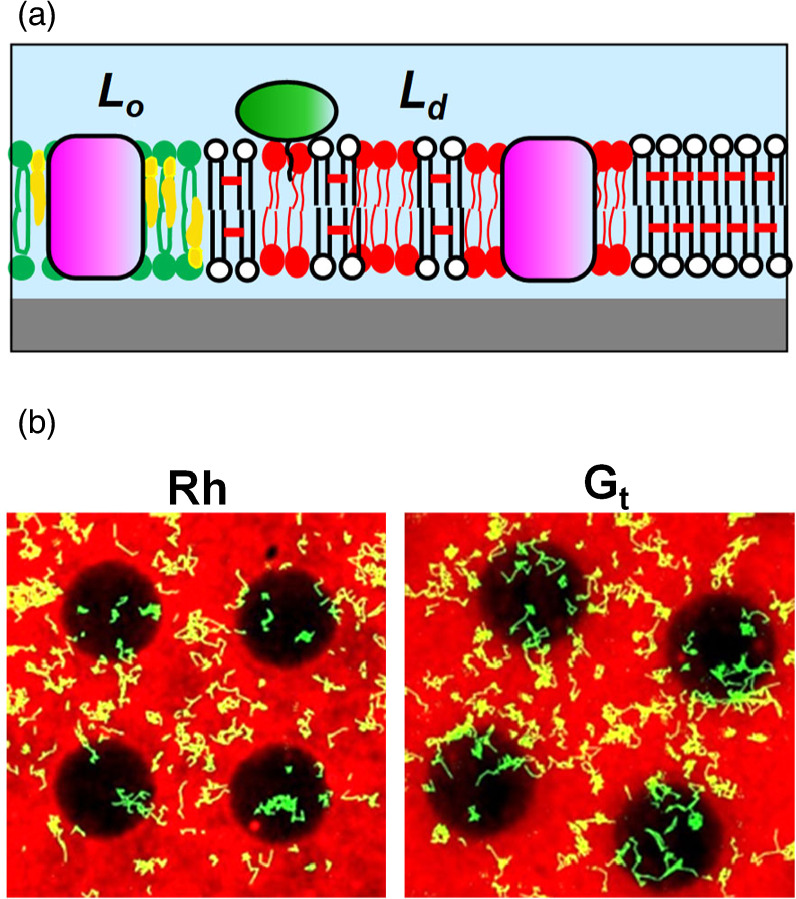
Membrane proteins play essential roles in the cell, including signal transduction and energy conversion, and they represent important targets of drugs. 26) We reconstituted a G protein coupled receptor (GPCR) of vertebrate retina (Rh) and its cognate G protein transducin (Gt) into a patterned membrane composed of Lo/Ld domains and observed their lateral distribution (Fig. 7). 11) We determined the partition and diffusion coefficients of Rh and Gt in the Lo-rich and Ld-rich regions. Both Rh and Gt were predominantly localized in the Ld phase, suggesting their low affinity to lipid rafts (raftophilicity). 11) The raftophilicity was presumably influenced by the lipid chains attached to proteins by the post-translational modifications. Measuring the raftophilicity for a variety of molecules in the signal transduction should reveal the functional significance of protein localization in the photosignal transduction.
Fig. 7. Reconstitution of membrane proteins: (a) schematic, (b) single-molecule-observation of Rh and Gt in a membrane with patterned phase separation. The pattern had circular areas of non-polymer (natural) region surrounded by composite (polymer/natural) region. The natural lipid bilayer (DOPC/DPPC/Chol (1:1:1) spontaneously separated into Lo phase (enriched with DPPC/Chol) and Ld phase (enriched with DOPC). The red fluorescence from TR-PE, which preferentially localizes in Ld phase, was observed predominantly in the composite region. The dark circular areas are enriched with Lo phase. The yellow lines are the trajectories of membrane proteins that diffused laterally in the membrane. Figures were modified from Y. Tanimoto et al., Biophys. J. 109, 2307 (2015) with permission of Cell Press.
Download figure:
Standard image High-resolution imageIn a different approach, we incorporated native biological membranes into the framework of polymeric bilayer. Thylakoid membranes purified from spinach leaves could be introduced into the corrals surrounded with polymeric bilayer together with synthetic phospholipid (DOPC) vesicles, spontaneously forming a laterally continuous and fluid 2D membrane. 12,27) Chlorophyll fluorescence arising from photosystem II (PSII) recovered after photobleaching, suggesting that the membrane components are laterally mobile. We confirmed the electron transfer activity of photosystem I (PSI) by observing the generation of NADPH in the presence of water-soluble ferredoxin and ferredoxin-NADP+ reductase. The lateral mobility of membrane-bound molecules and the functional reconstitution of major photosystems provide evidence that the hybrid thylakoid membranes could be an excellent experimental platform to study the 2D molecular organization and machinery of photosynthesis. In a different line of study, we directly transferred a mammalian GPCR, dopamine D2 receptor (D2R), from Chinese hamster ovary (CHO) cells into the patterned membrane using vesicles that bud off from the cell membrane (blebs). 28) D2R molecules were effectively reconstituted in a nanometer-sized thin cleft formed between a patterned SLB and PDMS. The density of D2R in the model membrane was high enough for the transient dimerization similar to the live plasma membrane. Although the mechanism of this enhanced reconstitution in the PDMS cleft is not fully understood, this finding points to a new possibility to use the PDMS cleft as a platform for reconstituting and studying membrane proteins under the quasi-physiological conditions.
3.3. Nanometer-sized gap between patterned membrane and PDMS
Although SLBs have a 2D membrane structure, the use of 3D micro-/nano-fluidics significantly enhances the utility of the model membrane. Therefore, it is important to establish a means to couple SLBs with 3D structures. We developed a methodology to chemically functionalize the surface of polymeric bilayer for attaching it with molecules, cells, and micro-fabricated structures (Fig. 3). 7) Polymeric bilayer with a reactive moiety was generated using a mixed bilayer of DiynePC and DiynePE. The amine group in DiynePE allowed to conjugate functional moieties (e.g. biotin) onto the membrane surface. Bonding PDMS and a polymeric bilayer with a molecular glue allows us to combine PDMS microstructures and substrate supported biomimetic membranes. We confined a membrane bound enzyme (cytochrome P450) with its water-soluble substrate in micro-compartments formed between PDMS and micro-patterned model membrane, and evaluated its functions. 7)
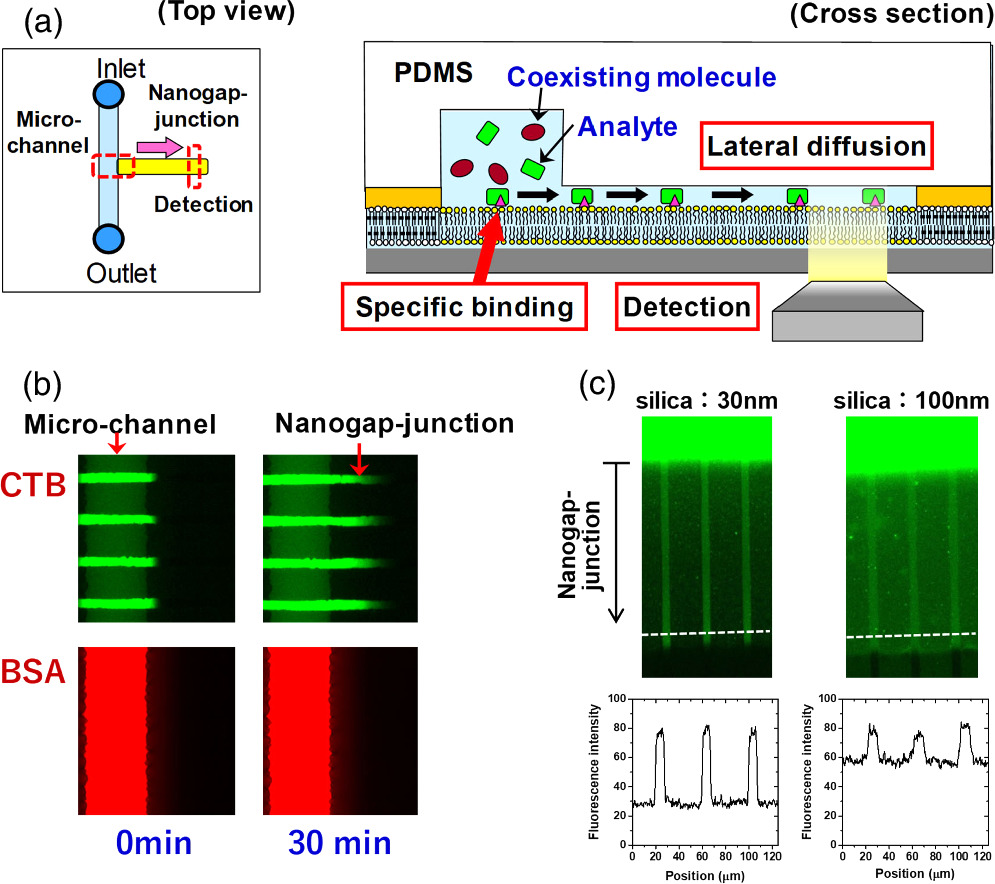
We combined the 2D patterned membrane and a nanometric detection volume by forming a very thin aqueous space between the fluid bilayer and a PDMS sheet (nanogap-junction) using an adhesion material with a pre-defined thickness (e.g. silica nanoparticles coated with hydrophilic polymer). The fabrication process of a nanogap-junction is outlined in Fig. 4. The adhesion material was attached onto the surface of the polymeric bilayer by the specific biotin-streptavidin linkage. The polymeric bilayer and the adhesion material were functionalized with biotin moiety. Then, streptavidin having four binding sites to biotin molecules was used to link them. In the same way, PDMS having the biotin-modified surface was attached onto the surface of the adhesion layer. The thickness of the nanogap-junction was determined by the adhesion layer, and we could generate a gap thickness of less than 100 nm. 29) The combination of a nanometric gap structure and a fluid lipid bilayer enabled to selectively transport and detect target molecules that could bind to the membrane surface by specific molecular recognition, whereas the nonspecific penetration of coexisting molecules was suppressed [Fig. 8(a)]. In Fig. 8(b), the model target molecule (cholera toxin subunit B: CTB) could bind to a fluid bilayer containing its ligand (GM1) and penetrate into the nanogap-junction by lateral diffusion (green fluorescence). In contrast, the model coexisting molecule (bovine serum albumin: BSA) could not bind to the membrane surface, and mostly remained in the solution in the microchannel. The selective transport of CTB into the nanogap-junction contributed to enhance the signal-background (S/B) ratio (i.e. the ratio of fluorescence intensities from specificcally bound CTB and the non-specific background noise measured in the polymeric bilayer regions). The use of silica nanoparticles as the adhesion layer enabled to control the thickness of the nanogap-junction accurately due to the uniform sizes of silica nanoparticles. 30) By using silica nanoparticles having a smaller size, we could generate a thinner nanogap-junction, which realized a higher S/B ratio [Fig. 8(c)]. In the micro-channel, the background fluorescence of BSA was very intense, and the fluorescence from membrane-bound CTB in the fluid bilayer was hardly observable. The signal from CTB was more clearly visible in the nanogap-junction due to the suppressed background fluorescence, because BSA molecules were mostly excluded. The background fluorescence intensity was more effectively suppressed for 30 nm nanoparticles compared with 100 nm nanoparticles, because a thinner gap is more effective in eliminating BSA, thus enhancing the S/B ratio even more drastically [Fig. 8(c)]. Furthermore, silica nanoparticles are mechanically robust, and the nanogap-junctions formed had a long life time (>week). The stability allows one to store a pre-assembled biochip having a nanogap-junction, thus eliminating the necessity for elaborate onsite assembly process. Compared to other techniques used for ultrasensitive detection, the nanogap-junction is unique in the following points. First, one does not need the total internal reflection of the incident light, significantly simplifying the illumination setup and enabling the use of cost-effective light sources, such as LEDs. Second, unlike zero-mode-waveguide, which measures single molecules at the bottom of nanoscopic wells, 31) a nanogap-junction offers an open 2D space where the lateral movement of single molecules can be tracked. It enables to monitor the lateral positions, movement, and binding/unbinding of membrane-bound molecules in real time. These features should provide a uniquely versatile platform for biosensing and analytical applications. Nanogap-junction can also be used for observing reconstituted membrane proteins. Single molecules of Rh could be observed in a nanogap-junction with a heightened S/B ratio compared with SLB facing a bulk aqueous solution. 32)
Fig. 8. Selective detection of target molecules that bind to the membrane and diffuse into the nanogap-junction: (a) schematic, (b) fluorescence microscopic observation of BSA-594 and CTB-488. CTB-488 bound specifically to GM1 in the membrane and penetrated into the nanogap-junction, whereas BSA-594 did not bind to the membrane, and mostly remained in the micro-channel. (c) The effect of silica nanoparticle sizes on suppression of background noise was assessed by introducing a mixture of BSA-488 and CTB-488 into the micro-channel. CTB-488 penetrated into the nanogap-junction, whereas BSA-488 mostly remained in the micro-channel. The background fluorescence arising from the nonspecific penetration of BSA-488 was lower in the nanogap-junction formed with 30 nm silica nanoparticles compared with the one formed with 100 nm silica nanoparticles. Figure 8 was modified from K. Ando, M. et al., Langmuir. 32, 7958 (2016) and M. Tanabe et al., Small. 14, 1802804 (2018) with permission of American Chemical Society and Wiley.
Download figure:
Standard image High-resolution image4. Conclusions
We have developed a patterned model membrane by lithographically polymerizing a synthetic lipid bilayer membrane and integrating it with natural lipid bilayers. The polymeric bilayer acts as a framework that defines the areas of natural lipid membranes. In addition, the embedded natural membranes are stabilized by the polymeric bilayer scaffold. The stabilization effect becomes more prominent as the pattern becomes smaller, because fluid lipid bilayers are surrounded and supported by the stable framework of polymeric bilayer. This assumption is supported by the fact that we can form a nanoscopic composite membrane of polymeric and fluid bilayers by adjusting the polymeric bilayer coverage. The patterned model membrane mimics the 2D organization in the biological membrane such as lipid rafts. By reconstituting membrane proteins, we can evaluate their physicochemical and functional properties in a controlled lipid membrane environment. We envisage to create a membrane circuit by rationally designing the membrane geometries and integrating membrane components including proteins and lipids. The membrane circuit should realize functional coupling of multiple membrane proteins that reproduce reaction cascades in the cell in a designed membrane geometry.
The patterned model membrane can also mimic the 3D architecture in the cell by attaching micro-/nano-structures to the polymeric bilayer surface. This technique enables to integrate the model membrane system with various analytical platforms. The integration, in turn, should enhance the unique advantages of model membranes, such as lateral mobility of molecules (fluidity) and suppression of non-specific protein adsorption. A nanoscopic space (nanogap-junction) was created by attaching a patterned membrane and PDMS with an adhesion layer having a defined thickness. The thin and small space with less than 100 nm thickness and 10 fl volume reproduces the environment existing in living cells. The combination of 2D and 3D membrane structures should become a platform of selective and sensitive biosensing, opening new avenues in biophysical studies as well as biomedical/food/environmental applications.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific research from Japan Society for the Promotion of Science (#26291031, #17H03666, #20H03225) and Iketani Science and Technology Foundation. Long-term collaboration and discussion with my colleagues and collaborators, including Dr. Fumio Hayashi, Dr. Shohei Maekawa, and Dr. Hiromasa Imaishi (Kobe University), are deeply appreciated.