Abstract
The dismal prognosis of pancreatic cancer has motivated research into identifying non-invasive 'liquid biopsy' biomarkers for early detection when treatment is most effective. Recently, exosomes—nanoscale membranous vesicles shed from tumour cells and which can be found circulating in the blood (and most bodily fluids)—have been discovered to contain a wealth of proteomic and genetic information, showing promise for pre-symptomatic screening and monitoring of disease. Here we examine recent findings highlighting the diagnostic value of exosomes in pancreatic cancer as well as the emerging use of lab-on-a-chip (LOC) technologies aiming to streamline exosome isolation and analysis.
Export citation and abstract BibTeX RIS
Circulating exosomes as novel biomarkers in PDAC
Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer and has the highest mortality rate of all major cancers, with median survival at <6 months and average 5 year survival rate at <4% in the United Kingdom [1, 2]. Major obstacles for improving clinical outcomes are diagnosis at advanced stage and limited treatment efficacy (resistance to chemotherapy and radiotherapy) [2]. Symptoms rarely develop with early-stage disease, which translates to more than 85% of patients presenting with surgically unresectable tumours [3]. Established risk factors for PDAC—such as chronic pancreatitis, obesity and tobacco smoking—are insufficient to risk-stratify the population [4]; furthermore, cross-sectional imaging (CT and MRI scans) can be prohibitively expensive for routine screening and are unable to detect potentially curable tumours with size of <1 cm in the context of early-stage or minimally-residual disease [5]. The non-biopsy blood test using the clinical standard serum biomarker carbohydrate antigen 19-9 (CA19-9) does not have the requisite specificity (82–90%) or sensitivity (79–81%) or even the capability to discriminate benign disease (e.g. pancreatitis) from overt pancreatic malignancy [6]. However, studies suggest that often a decade or more lapses before the founder malignant clone in the pancreas (early panIN lesions) becomes metastastic PDAC, suggesting a window of opportunity for early detection [7].
Tumour-derived exosomes are attracting increasing interest in the liquid biopsy field on the premise that circulating exosomal cargo may include novel markers for blood-based screening, diagnosis and follow-up of human cancer, including pancreatic cancer [8]. Exosomes, are nanometre-sized (30–150 nm) vesicles released by most cells via exocytosis during fusion of multivesicular bodies (MVEs) with the plasma membrane [9]. Once considered as cellular debris, exosomes are now recognized as promising tumour surrogates because they deliver enriched biomarkers in the form of proteins, RNAs and DNA [10, 11]. Through the circulation, exosomes act as functional mediators of neighbouring or long-distance cell–cell communication regulating a variety of physiological and pathological processes in a hormonal manner [10]. They play a critical role in human cancer, contributing to tumourigenesis and metastasis by modulating tumour immune responses and priming distant organs for metastatic dissemination of tumour cells [12–16]. Exosomes possess distinct advantages as biomarkers, including non-invasive collection, high abundance in biofluids (~105 ml−1) with their number increasing further in cancer patients [17, 18] as well as stability over time and inclusion of proteins and nucleic acids from the parental tumour. Moreover, in the setting of early-stage disease, living-cell secreted exosomes can be detected potentially earlier in the bloodstream than necrosis-caused release of cell-free DNA (cf-DNA) or circulating tumour cells (CTCs), the latter two usually arising at more advanced tumour stages [19, 20].
Identifying exosome-associated biomarkers for early detection is challenging due to bottlenecks relating to lack of standardised methods for exosome isolation and characterisation. As most cells secrete exosomes, identifying 'cancer-specific' exosome signatures via proteomic and genomic approaches will go a long way in providing reliable biomarkers for disease diagnostics; the past few years have seen some very promising growth for the exosome field.
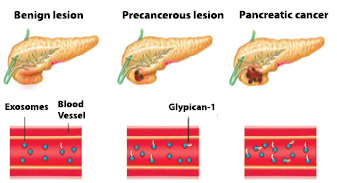
In a recent seminal study, Melo and colleagues [21] used proteomics to identify glypican-1 (GPC1) as a membrane-bound protein preferentially detectable in cancer-associated exosomes secreted from pancreatic and breast cancer cell lines. The group demonstrated increased levels of GPC1+ circulating exosomes in mice and humans with pancreatic and breast cancer. Strikingly, they also identified exo-GPC1 to scale with tumour stage in a concentration-dependent manner and found that exo-GPC1 levels were the highest in patients with pancreatic cancer and pancreatic cancer precursor lesions (IPMNs, intraductal papillary mucinous neoplasms) than in healthy controls or benign pancreatic diseases or chronic pancreatitis. Remarkably, ROC curves, which plot sensitivity versus (1-specificity), demonstrated an area under the curve (AUC) of 1.0 suggesting discrimination with 100% sensitivity and specificity of pancreatic cancer at all stages from non-pancreatic cancer controls. Interestingly, direct ELISA analysis of serum GPC1 showed a much lower sensitivity (~82%) and specificity (75%) compared to exo-GPC1 indicating the need to measure GPC1+ exosomes rather than free circulating GPC1 to achieve optimal discrimination. Longitudinal observations of PDAC patients showed serum exo-GPC1 levels to be responsive to surgical intervention, with removal of the tumour leading to a significant reduction in exo-GPC1, while also serving as an independent prognostic and predictive marker for disease-specific survival. Furthermore, in mice developing pancreatic cancer, levels of GPC1+ exosomes were increased early in tumourigenesis (panIN stage), even before the appearance of MRI detectable pancreatic masses, with Exo-GPC1 levels further increasing proportionally with time (and tumour burden) and severity of disease (histopathology). This was a study that reverberated across the cancer community, with the main caveat being the low sample size for the early-stage disease group (5 cases for IPMNs) in the discovery cohort, highlighting an issue relating to biospecimen availability and underscoring a need for biospecimen collection before resection from patients with early-stage pancreatic malignancy. With independent confirmation of its performance from other laboratories, GPC1 in circulating exosomes may soon be established as a more reliable biomarker for diagnosis and prognosis of pancreatic cancer than the currently FDA-approved serum biomarker CA-19-9 (figure 1).
Figure 1. Exosome glypican-1 as a non-invasive biomarker for pancreatic cancer. Exosome glypican-1 has been found according to a study by Melo et al to have 100% specificity and sensitivity as a 'liquid biopsy' marker for detecting early stage pancreatic cancer. Concentration of exosome glypican-1 in serum positively correlates with disease progression and can discriminate healthy/benign conditions from early stage and late-stage pancreatic cancer.
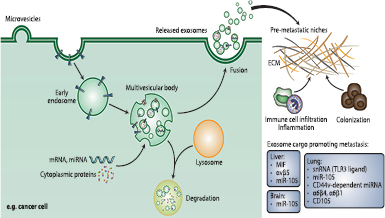
Download figure:
Standard image High-resolution imageA recent international collaborative initiative led by Lyden and colleagues [13] reported that tumour-secreted exosomes precondition specific organs for metastatic invasion long before cancer cells reach these organs, supporting the century-old 'seed and soil' theory of metastasis. The authors isolated exosomes from organotropic human and mouse breast and pancreatic cancer cell lines known to preferentially metastasise to the lung and liver respectively. Then, they injected these fluorescently labelled exosomes into nude mice and examined their biodistribution and uptake in distant organs after 24 h by NIR-whole lung imaging and confocal microscopy. Indeed, they observed that exosomes from the different cancer models selectively interacted with the same future metastatic organs as their cell of origin. To test exosome-guided education of target organs functionally, the group injected luciferase-expressing cancer cells via the tail vein or intracardially, with capacity to metastasise to either lung (4175-LuT) or bone (1833-BoT) in mice previously educated with either lung-tropic or bone-tropic tumour exosomes. Education with lung-tropic exosomes not only increased the lung metastastic capacity of 4175-LuT cells but surprisingly also increased the lung metastatic capacity of 1833-BoT cells. These data suggested that tumour exosomes prepare pre-metastatic niches to facilitate organ-specific metastasis and redirect metastatic distribution even for cancer cells poorly capable of metastasising to these sites. Proteomic analysis identified distinct exosomal integrin patterns as key determinants of organotropic metastasis to the lung, liver and brain. In particular, lung-tropic exosomes highly expressed integrins α6β4 and α6β1 and found to home in laminin-rich lung microenvironments to interact with lung fibroblasts and epithelial cells, while PDAC-derived liver-tropic exosomes had upregulated expression of integrin ανβ5 which directed them in fibronectin-rich liver niches to intereact with liver Kuppfer cells. Blocking these exosomal integrins via short hairpin markedly reduced lung or liver metastasis. Uptake of integrin-expressing tumour exosomes by organ-specific resident cells activated Src phosphorylation and proinflammatory S100 gene expression to make the organ-specific microenvironment favourable for future metastasis. Importantly, their clinical data also showed that exosomal integrin β4 is higher in breast cancer patients with lung metastasis and elevated exosomal integrin αν in pancreatic cancer patients who developed liver metastasis than in those without metastasis. These results indicate that exosomal integrin signatures may be used as organotropism biomarkers to predict organ-specific metastasis in cancer patients or as potential candidates for anti-metastastic therapy.
Another study conducted by Costa-Silva et al reported a functional role for PDAC-derived exosomes in facilitating metastatic dissemination through stromal re-education [22]. Exosomes released from pancreatic cancer cell lines were found to overexpress the macrophage migration inhibitory factor (MIF). Using fluorescent labelling and tracking, MIF+ PDAC-derived exosomes were retrorbitally injected into mice and found to be taken up by Kupffer cells in the mouse liver triggering a cascade of events—TGF-β release from Kuppfer cells, fibronectin deposition from resident hepatic stellate cells and recruitment of bone-marrow derived cells to fibronectin-enriched hepatic sites—ultimately leading to formation of pre-metastatic niches in the liver making the local environment more hospitable to residence of circulating metastatic cancer cells. Notably, circulating MIF+ exosomes were detectable in mouse models before invasive disease was identified in the pancreas, suggesting that priming of favourable metastatic niches in the liver may occur with premalignant lesions of the pancreas. MIF concentration in circulating exosomes was also higher at diagnosis among patients that developed recurrence of disease at 5 years than those who showed no such evidence. Intriguingly, MIF blockade could prevent liver metastases in mouse models, suggesting that tumour exosome contents may prove to be clinically relevant for both prognosis as well as the development of novel targeted therapies (figure 2).
Figure 2. Exosomal cargo can direct and predict organotropic metastasis. Exosomes are extracellular vesicles (30–150 nm) that are released from cells upon fusion of an intermediate endocytic compartment, the multivesicular body (MVB), with the plasma membrane. This releases intraluminal vesicles into the extracellular milieu and the vesicles thereby are what we know as exosomes. The MVB can alternatively deliver content to lysosomes for degradation. Both amount and molecular composition of exosomes depend on the state of the parental cell. Exosomes have pleiotropic physiological and pathological functions with an emerging role in cancer metastasis by establishing pre-metastastic niches in distant organs and directing as well as predicting organotropic metastasis [13, 22–28]. Exosome-based liquid biopsies could potentially be used to assess metastatic propensity in certain cancers and guide follow-on treatment.
Download figure:
Standard image High-resolution imageAn independent case-control study by Que et al examining PDAC-associated exosomal miRNAs found moderate discrimination of cases from control by exo-miR-21 and exo-miR17-5p [29]. Another study conducted by Madhavan and colleagues reported that a combination of five proteins (CD44v6, Tspan8, EpCAM, MET and CD104) and four miRNAs (miR-1246, miR-4644, miR-3976 and miR-4306) in circulating tumour exosomes could distinguish pancreatic cancer cases from non-cases, which included healthy control, chronic pancreatitis patients and individuals with benign pancreatic disease, but excluded non-pancreas-related malignancies, with 93% specificity [30].
Another interesting study by San Lucas and colleagues showed that circulating exosomes from PDAC patients contained a large proportion of tumour DNA and RNA [31]. Next-generation sequencing of the nucleic acid cargo contained within exosomed revealed a wide number of relevant cancer biomarkers, including copy number profiles, point mutations, insertions and deletions, gene fusions and mutational signatures.
In summary, circulating tumour exosomes should be considered on the short list of biomarkers with tremendous potential for early detection of pancreatic cancer (table 1). Liquid biopsies using shed exosomes has the potential to be used as a clinical tool for early-stage cancer diagnosis, therapeutic stratification and treatment monitoring, precluding the need for direct tumour sampling.
Table 1. PDAC-associated exosome biomarkers.
| Exosomal marker | Diagnostic value | Specificity | Sensitivity | Sample size | Study design | Study year |
|---|---|---|---|---|---|---|
| Glypican-1 | Discriminates healthy subjects versus benign disease versus pre-cursor lesions versus PDAC | 100% | 100% | Healthy (n = 100), benign (n = 26), precursor (n = 5), PDAC (n = 190) | Case-control | Melo et al [21] |
| Integrin ανβ5 | Predicts organotropic metastasis | NA | NA | Healthy (n = 13), PDAC no relapse (n = 14), liver metastasis (n = 13) | Case control | Hoshino et al [13] |
| Macrophage migration inhibition factor (MIF) | Predicts liver metastasis and recurrence | NA | NA | Healthy (n = 15), liver metastasis (n = 18), disease progression (n = 12), no disease progression (n = 10) | Case control | Costa-Silva et al [22] |
| miR-21 and miR-17-5p | Discriminates PDAC versus healthy controls | 81.5% and 92.6% | 95.5% and 72.7% | Healthy (n = 8), PDAC (n = 22), benign (n = 6) | Case control | Que et al [29] |
| miR-1246, miR-4644, miR-3976, miR-4306 and CD44v6, Tspan8, EpCAM, MET and CD104z | Discriminates PDAC versus healthy and benign conditions | 80% | 100% | Pancreatic malignancies (n = 131), non-pancreatic malignancies (n = 12), benign (n = 22) | Case control | Madhavan et al [30] |
| DNA and RNA (copy numbers, gene fusions, mutational spectrum) | Molecular characterization of tumour for guiding treatment | NA | NA | 3 patients with pancreaticobiliary cancers | Case | San Lucas et al [31] |
Conventional methods for exosome isolation
Despite their enormous potential in disease diagnostics and cancer research, exosome isolation and recovery from biofluids remains technically challenging thereby limiting progress in basic exosome research as well as clinical utilisation. This is largely due to their nanoscale size (30–150 nm). Conventional techniques separate exosomes based on their size and buoyant density [32].
The most common method for isolating exosomes from biofluids relies on ultracentrifugation—a size-dependent method of separation that includes differential centrifugation steps reaching speeds of of up to 200 000 × g to pellet exosomes from supernatant [33, 34]. However, this is a labour-intensive, time-consuming procedure that requires specialist laboratory equipment not typically available in routine clinical settings or hospital laboratories and is also highly inefficient with regard to exosome yield (5–25%) and purity [32, 34]. Variations on this method, such as adding a sucrose-gradient centrifugation step can increase purity and recovery rate. Density-gradient centrifugation is an equilibrium method, where a sample is spun in a tube that contains a density gradient of a viscous solution (usually iodixanol), such that bioparticles separate based on their buoyant density or isopycnic point. This technique has some of the same problems with conventional ultracentrifugation, and requires the same expensive, bulky equipment making it impractical for routine use and clinical applications. Size overlap with other membranous vesicles, such as shed microvesicles, apoptotic blebs or large protein aggregates means that in order to isolate a pure population of exosomes a combination of techniques is necessary based on both physical (e.g. size, density) and biochemical parameters (e.g. presence of certain exosome-protein markers involved in their biogenesis).
Conventional methods for exosome characterisation
Molecular characterisation of isolated exosomes is typically carried out using standard techniques, such as western blot, ELISA and mass spectrometry; all lengthy processes that require large sample volumes and concentrated exosome samples as the presence of contaminants can bias the information obtained from these assays [32]. During flow cytometry, the workhorse technique for high-throughput analysis of cells, each exosome passes individually through a laser spot and its emitted scattered and fluorescent light is measured. However, flow cytometry has demonstrated limited reliability for detecting particles smaller than 200 nm. Thus, exosomes are usually tagged with antibody-coupled bead prior to analysis in order to increase detection sensitivity. Nanoparticle tracking analysis (NTA) is another commonly used technique that can detect single exosomes. NTA enables quantitative sizing of exosomes by combining light microscopy and software that analyses the particles' Brownian motion [35]. The software tracks the movement of exosomes as they diffuse through the field-of-view and calculates the diameter of the particle based on its rate of Brownian motion. The Brownian motion can be related to particle size using the Stokes–Einstein relationship, which requires knowledge of only the temperature and the viscosity of the suspending fluid. NTA used in light scattering mode allows sizing of exosomes while the fluorescence mode can be used for profiling a labelled molecular exosome marker. Another technique that has been used to detect and characterise single exosomes is transmission electron microscopy (TEM). The diameter of exosomes is smaller than optical wavelengths which means exosomes cannot be seen under a conventional light microscope. Using the short wavelength of electrons, TEM can resolve individual exosomes and provide information regarding size, concentration and analysis of surface markers, especially when combined with immunogold labelling. However, the extensive sample processing (dehydration, fixation and metallisation) can be limiting. Atomic force microscopy (AFM) has also been used to characterise single exosomes but the inherent low-throughput can also be problematic.
There is a pressing need for efficient and cost-effective methods for large-scale purification and analysis in clinical settings that are standardised across laboratories to allow clinical implementation. Microfluidic lab-on-a chip approaches can effectively address many of the technical hurdles surrounding exosome isolation and analysis owing to their inherent advantages in miniaturisation, integration, automation and parallelisation. We expect the application of micro- and nanotechnologies to streamline exosome purification and detection and pave the way for novel point-of-care diagnostic devices for exosome-based liquid biopsies. Below we spotlight some of the state-of-the-art microfluidic/lab-on-chip devices for isolation and detection of exosomes.
Lab-on-a-chip technologies for exosome isolation
Over the last few years, novel approaches have been implemented to scale microfluidic techniques to the nanoscale for their application to exosome isolation. There are inherent challenges to this scaling due to the susceptibility of nanoscale channels to clogging, the expense of nanoscale lithography and unfavourable scaling of forces acting on nanoscale objects.
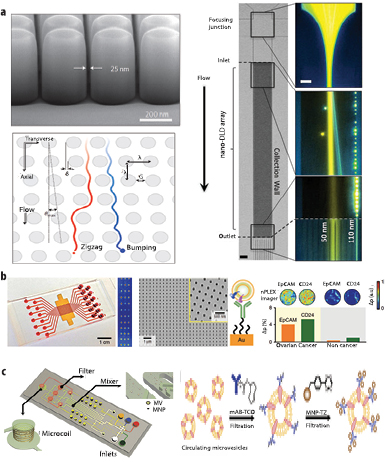
IBM in collaboration with Icahn School of Medicine has recently developed a nanofluidic chip that can achieve size-based separation of bioparticles down to 20 nanometres—a scale particularly relevant for separating exosomes [36]. The nanofluidic chip makes use of a technique known as nanoscale-deterministic lateral displacement (nano-DLD), where a liquid sample is passed, in continuous flow through an array of asymmetric nanoscale pillars separating particles by size down to tens of nanometers resolution. DLD has been previously applied to separate microparticles (e.g parasites, and circulating tumour cells) but in this study the authors demonstrated the same principle can be modified to separate nanoscale particles in spite of the diffusion, a hallmark of particle dynamics at these small scales. The nano-DLD array could deflect larger particles while allowing smaller particles to flow through the gaps of the pillar array unabated. Furthermore, this is a passive, label-free technique without the need of using antibodies. Fabrication of the nano-DLD pillars, however, required the use of e-beam nanolithography, which although capable of very high resolution, can also be a very expensive manufacturing process and considerably slower to optical lithography. Also, the presence of nanoscale features of the device can increase the chance of clogging. Finally, the volumetric flow rate—and thus the overall throughput of the device, at the tested input pressure of 200–800 kPa was rather low at 0.1–0.2 nl min−1 yielding 104–106 particles per minute.
Another creative approach by the Liu group at University of Texas where they developed a microfluidic device consisting of ciliated micropillars with a hierarchical structure using conventional microfabrication techniques [37]. Using silicon photolithography, electroplating and metal-assisted etching, they fabricated micropillars with silicon nanowires etched on the sidewalls of the micropillars that could physically trap exosomes in the range of 40–100 nm, while simultaneously filtering out proteins and cellular debris. They further demonstrated that trapped exosomes could be recovered intact by dissolving the porous nanowires in PBS buffer. The device was tested with a solution containing 83 nm, 120 nm and 500 nm lipid vesicles and nanoparticles. The highest trapping efficiency (60%) was obtained for 83 nm vesicles, followed by 120 nm vesicles (45%), while the retention of 500 nm nanoparticles was only 10%.
Davies et al prepared a microfluidic filtration system using in situ prepared nanoporous membranes to size-selectively isolate exosomes from unprocessed whole blood in a single step [38]. The filtering membranes consisted of photopatterned porous polymer monoliths integrated into poly(methyl methacrylate) microfluidic chips; the porous properties of the membrane could be adjusted acting as a one-way tunable size-cut off filter (<500 nm) for exosome and extracellular vesicle isolation. The sample was driven either via pressure or DC electrophoresis. The authors noted that DC electrophoresis led to higher purity of extracted exosomes with proteins being less affected by electric field compared to phospholipid vesicles. However, the main drawback of forced filtration or other physical trapping approaches is clogging of the device and/or potential damage to the exosomes.
Immunoaffinity-based isolation strategies have also been implemented into microfluidic devices either via functionalised microchannel surfaces with antibodies targeting exosomal markers or by incorporating antibody-coupled particles and/or magnetic beads. Kanwar et al developed ExoChip a microfluidic chip that utilized anti-CD63 functionalized circular microchambers for immunoaffinity based capture of exosomes from blood serum with subsequent fluorescent staining using the lipophilic dye (DiO) for quantification [39]. To enhance the available surface area for antibody immobilization manipulation of immunomagnetic beads in a microfluidics is a viable alternative. Zhao et al developed ExoSearch, a microfluidic, multi-stage circuit platform, which combined on-chip continuous flow-mixing and immunomagnetic isolation with an in situ multiplexed exosome immunoassay [40].
Dudani et al also reported a microfluidic device that makes use of intertial forces that naturally arise in confined microchannels at low Reynolds numbers. The device used a high aspect ratio microchannel, which generates an inertial lift force that forces flowing particles on the centerline of the channel [41]. However, premixing and incubation of capture beads with samples were needed with this approach. The Trau group also developed a novel method, known as nanoshearing [42] that relies on electrohydrodynamic lateral fluid flow generated within a few nanometers of an electrode surface to enhance the efficiency of immunoaffinity-based capture of exosomes by increasing the number of exosome-antibody (surface-bound) collisions, as well as reduce the non-specific adsorption of weakly bound molecules.
The group of Lee et al used an acoustophoretic microfluidic system to isolate microvesicles from cell culture supernatant and blood plasma. Acoustophoresis, which relies on the generation of surface standing acoustic waves, achieved size-dependent separation of microvesicles from other blood components. The technique had reasonably good throughput (0.4–0.7 µl min−1), no clogging and high exosome recovery rates >80%.
In the future, label-free active separation techniques that utilize non-inertial forces—dielectrophoretic, acoustophoretic [43] or oprtical forces- to sort nanoscale bioparticles based on physical characteristics (e.g. size, density, compressibility) may provide sensitive, high-throughput, clogging-free devices to isolate nano-sized exosomes in a gentle non-contact manner (table 2).
Table 2. Microfluidic platform technologies for exosome isolation.
| Microfluidic isolation technology | Sample | Isolation yield | Throughput | Label-free isolation | Isolation capacity | Study year |
|---|---|---|---|---|---|---|
| Nanoscale deterministic lateral displacement (nano-DLD) | Urine sample | 104–106 particles min−1 | 0.1–0.2 nl min−1 | Yes (based on size) | Continuous flow | Wunsch et al [36] |
| Ciliated micropillars | Liposome solution | 45–60% | 10 µl min−1 | Yes (based on size) | Saturation limit 100 µl | Wang et al [37] |
| Microfluidic filtration with photopatterned nanoporous membranes | Whole blood | ~3% | 1 µl min−1 | Yes (based on size) | Saturation limit 100 µl | Davies et al [38] |
| Immunoaffinity-based capture in circular microchambers (ExoChip) | Serum | 15–18µg in 60 min | 8 µl min−1 | No | ~400 µl | Kanwar et al [39] |
| Acoustohporetic isolation | Culture media and packed RBC units | 80% | 0.4–0.7 µl min−1 | Yes (based on size) | Continuous flow | Lee et al [43] |
| Online immunomagnetic capture (ExoSearch) | Plasma | 42–97.3% | 10 µl min−1 | No | Continuous flow (10 µl to 10 ml) | Zhao et al [40] |
| Bead affinity capture with inertial solution exchange | Culture media | NA | 140 µl min−1 | No | Continuous flow | Dudani et al [41] |
Lab-on-a-chip technologies for exosome detection
Several innovative microfluidic approaches have also been implemented to enhance exosome detection sensitivity. Surface plasmon resonance (SPR) biosensors have garnered a lot of attention in recent years due to their exceptional sensitivity in detecting an exceedingly small number of analytes in a sample. In SPR, the region of detection sensitivity extends only a few hundred nanometers above the surface and is thus well suited for ultransensitive detection of nanoscale exosomes. Im et al developed a nanoplamonic exosome sensor chip (nPLEX) that comprises periodic arrays of nanoholes patterned in a gold film within parallel microfluidic channels to detect exosomal protein markers in a multiplexed, high-throughput and label-free manner [44]. The nPLEX sensor relied on transmission SPR enabling real-time monitoring of exosome binding events and offering improved sensitivity over conventional ELISA and flow cytometry. By combining these nanohole arrays with a miniaturised imaging setup, Im et al demonstrated that the nPLEX sensor chip can be scaled for massively parallel measurements—105 independent nanohole arrays, each functionalised with a different affinity ligand. The nPLEX device could readily differentiate samples from ovarian cancer and healthy controls by detecting exosome-associated molecular signatures (CD24 and EpCAM) requiring sample volumes as little as 0.3 µl.
Another creative approach coming from Shao and colleagues where they developed a miniaturized nuclear magnetic resonance based microfluidic platform (µNMR) to sensitively measure exosomes [45]. In this system, exosomes were first labeled with magnetic nanoparticles (MNP) targeted to specific exosomal protein markers. These exosomes were then captured onto a membrane filter in a size-dependent manner and subsequently washed to remove unbound beads. In-line µNMR detection of the surrounding H2 molecules was then performed, and the change in T2 relaxation signal due to the presence of MNPs in the detection region, was used to profile the exosomes. The microfluidic µNMR chip was 104-, 103- and 102 more sensitive than western blotting, ELISA and NTA respectively for detecting and quantifying exosomal biomarkers. Following up, the group demonstrated clinical utility using the device to differentiate glioblastoma multiform (GBM) microvesicles from non- tumour host-cell derived microvesicles (figure 3).
Figure 3. Lab-on-a-chip microfluidic approaches towards exosome isolation and analysis. (a) IBM's nano-DLD chip [36] (nanoscale deterministic lateral displacement) can achieve passive, label-free, size-dependent separation of exosomes between 20 and 110 nm with sharp resolution. The chip's nanofabricated pillar array causes differently sized bioparticles to follow differential trajectories (zigzag versus bumping) enabling highly sensitive fractionation of exosomes according to their size. (b) The n-PLEX chip [44] utilises transmission-based surface plasmon resonance (SPR) to detect exosomes that are selectively captured on gold nanohole sensor arrays (<200 nm probing depth) functionalised with antibodies against exosomal markers. The n-PLEX chip can be integrated with a multichannel microfluidic cell for high throughput and parallel detection. The device is further integrated with a CMOS imaging setup to monitor changes in the intensity of transmitted light and was validated by detecting exosomal markers (EpCAM and CD24) in ovarian cancer samples. (c) The µNMR chip [45] was designed to detect magnetic nanoparticles (MNP)-targeted exosomes, concentrate MNP-tagged exosomes (while removing unbound MNPs) and provide in-line µ-NMR detection. Panel (a) reprinted by permission from Macmillan Publishers Ltd: [36], copyright (2016). Panel (b) reprinted by permission from Macmillan Publishers Ltd: [44], copyright (2014). Panel (c) reprinted by permission from Macmillan Publishers Ltd: [45], copyright (2012).
Download figure:
Standard image High-resolution imageIn another effort for improving exosome detection, Zhang et al fabricated a graphene oxide/polydopamine (GO/PDA) nanointerface [46]. The GO-induced formation of a 3D nanoporous PDA surface coating enabled highly sensitive on-chip immunoaffinity-based capture of exosomes while minimizing non-specific adsorption by polyethylene glycol (PEG). The group from Lee et al developed an integrated magnetic-electrochemical exosome detection platform (iMEX). Magnetic beads were used for immunoaffinity-based exosome capture and labeling; captured exosomes were then detected via electrochemical sensing which offered high sensitivity through signal amplification with redox-active reporters. The readout measured electrical currents and the whole device was packaged into a compact, hand-held, low-power device. iMEX further provided fast (<1 h), sensitive (104 exosomes/ µl detect · limit) and parallelized measurements, and was validated by profiling exosome samples collected from ovarian cancer patients [47].
Yoshioka et al developed ExoScreen 96-well plate for sensitive liquid biopsy of exosomes offering rapidity, lower sample volumes and higher sensitivity compared to conventional ELISA [48].
The Cleland group developed a nanoparticle analyzer for high throughput, label-free quantitative counting and sizing of synthetic or biological nanoparticles, including exosomes, with single-particle resolution [49]. The analyzer consisted of a microfluidic-based nanopore device that integrated electrical and fluidic components: The fluidic channel consisted of a primary nanoconstriction for particle detection and a fluidic restriction that provided a balancing electrical resistance. Together, these components form a fluidic voltage divider that yielded wide bandwidth electrical detection of particles as they passed through the nanoconstriction. Constant voltage is applied to bias electrodes, generating an ionic electrical current in the analyte, with voltage drops across the fluidic resistor and nanoconstriction. A sensing electrode is embedded via optical lithography in the channel between the fluidic resistor and the nanoconstriction. When a particle enters the constriction, it alters the ionic electrical current, and because of the voltage division between the fluidic resistor and nanoconstriction, changes the electrical potential of the fluid in contact with the sensing electrode. The voltage signal readout ΔVout from the sensor electrode scales with particle diameter and is proportional to particle diameter allowing particle sizing even for very rapid transit times of nanoparticle through the nanopore(<3 µs) with excellent signal-to-noise ratio.
The group by Chang et al developed a 2-part microfluidic platform for simultaneous exosome lysis and exosomal RNA detection [50]. First on-chip acoustic lysising was achieved via high-frequency surface acoustic waves (SAW) generated by a single interdigital transducer on a piezoelectric substrate and focused in a microfluidic chamber containing an exosome sample. The refraction of SAW into the liquid caused an acoustic streaming force which ruptured the exosome membrane releasing the intravesicular contents for downstream RNA analysis in an ion-exchange nanomembrane sensor; the sensor consisted of two reservoirs separated by a membrane. RNA detection in the sensing reservoir occurs via hybridization of complementary oligos immobilized on the surface of the membrane to exosomal RNA. In the future improved nano-based field effect transistors and improved biosensor designs may further improve exosomal diagnosis (table 3).
Table 3. Microfluidic platforms for exosome detection.
| Microfluidic detection technology | Detection sensitivity | Label-free detection | Multiplexing capability | Readout | Study year |
|---|---|---|---|---|---|
| Nanoplasmonic detection (nPLEX) | Very high—3000 exosomes (670 aM) | Yes | Yes | Optical–CMOS based (spectral shift) | Im et al [44] |
| µNMR chip | Ηigh—10 000 exosomes | No | Yes | µNMR signal (T2 relaxation time) | Shao et al [45] |
| Graphene oxide/polydopamine (GO/PDA) nanointerface | Very high—80 aM | Yes | No | Fluorescent | Zhang et al [46] |
| Integrated magnet-electrochemical detection platform (iMEX) | Moderate—< 105 exosomes | No | Yes | Electrical | Jeong et al [47] |
| ExoScreen | ELISA-grade | No | No | Fluorescent (plate reader) | Yoshioka et al [48] |
| Nanoparticle analyser (nanopore device) | Single particle (counting and sizing) | Yes | Yes | Electrical | Fraikin et al [49] |
| Acoustic exosome lysing and exo-RNA detection in ion-exchange nanomembrane sensor | ~2 pM | — | Yes | Electrical | Taller et al [50] |
Concluding remarks
Non-invasive liquid biopsies (circulating exosomes, circulating nucleic acids and circulating tumour cells) are promising to revolutionise the cancer diagnostic landscape by providing a more effective and suitable alternative to the gold standard tissue biopsy for cancer screening, longitudinal monitoring of the disease as well as guiding treatment decisions by informing clinicians on the evolving mutational landscape of the tumour. In the context of pancreatic cancer circulating exosomes have shown tremendous potential for early-stage diagnosis that could in principle facilitate curative surgical therapy.
However, the clinical utility of exosomes in the managemet of PDAC is still too early and independent confirmation of the performance of promising exosomal biomarkers (e.g. glypican-1) is needed with larger cohorts for pre-cursor lesions. Given the scarcity of human PanIN/IPMN biospecimens, this will likely necessitate international collaborations of clinical centres and universities. Early-stage PanIN lesions are common with increasing age and rarely progress into PDAC [51]. On the other hand, late-stage PanIN lesions carry a higher risk of progressing into invasive disease. Thus detection of early-stage disease without reference to the specific PanIN stage and/or the likelihood of its progression to PDAC will inherently lack specificity for life-threatening invasive PDAC. Clinical trials with hard end points will also be needed to evaluate if early-stage intervention leads to better clinical outcomes.
In parallel, microfluidic-based technologies will continue to play an instrumental role in improving exosome-based liquid biopsies in the future. We expect that microfluidic approaches that combine both physical and biochemical filters for integrated on-chip purification and detection while avoiding sample pre-processing or moving parts will be the way forward. Portability, cost-effectiveness, high-throughput and multiplexing capability, scalability, minimal sample volume consumption and ease of fabrication are all factors to be considered for the next-generation microfluidic-based devices for exosome liquid biopsies. The liquid biopsy market is still in its infancy and largely unsaturated providing a wide-open IP space. Innovative physical and engineering approaches to develop micro and nanoscale technologies are promising both in terms of commercialization as well as in expediting and streamlining basic exosome research and workflow.
Acknowledgments
This work was supported by the European Research Council (grant agreement 282051). TJL is supported by the Whitaker International Program and James Dyson Foundation