Abstract
Pyronin Y is an environment-sensitive probe which labels all double-stranded RNA in live cells. Methods to determine which RNA species Pyronin Y may be labeling are limited due to the lack of studies aimed at determining whether this probe has different spectroscopic properties when bound to specific transcripts. A major issue is that transcripts are difficult to isolate and study individually. We detected transcripts directly in their biological environment allowing us to identify RNA species on the basis of their location in the cell. We show that the phasor approach to lifetime analysis has the sensitivity to determine at least six different RNA species in live fibroblast cells. The detected lifetime differences were consistent among cells. To our knowledge this is the first application of a spectroscopic technique aimed at identifying Pyronin Y labeled RNA subtypes in living cells.
Export citation and abstract BibTeX RIS
1. Introduction
The ability to visualize transcript dynamics in living cells may aid in understanding RNA properties including intron processing, localization, redirection and transport and in monitoring these cell developments as a function of time (Bao et al 2009, Santangelo et al 2006, 2005, 2004). To observe the natural dynamics of RNA in live cultures, the inducible properties of a probe are often used to provoke indirect fluorescence of a transcript and enable its visualization in living cells. The application of different probes has enabled the identification of unique RNA properties including rearrangement of messenger ribonucleoproteins (mRNPs) upon interaction with a nuclear pore complex to initiate export (Grunwald et al 2011), identification of the mobility mechanisms undertaken by transcripts including GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and k-ras (Santangelo et al 2004) messenger RNA (mRNA) as well as the redirection of β-actin mRNA during cell development (Dahm and Kiebler 2005).
The ability of a fluorophore to surpass partitioned regions within living cells including membrane barriers is pivotal for the labeling of desired transcripts. Recently, dual molecular beacons have become a common application to target a single transcript species and induce fluorescence often through the application of FRET (fluorescence resonance energy transfer) (Santangelo et al 2006, 2005, 2004). Although advantageous in their specificity, molecular beacons often undergo undesired sequestering in nuclear regions. Chemical alteration of the beacon or the addition of a transfer RNA (tRNA) to the molecular structure could result in complete nuclear exclusion of the beacon with or without the attached transcript (Chen et al 2007, 2008). Targeting the transcript with large proteins may inhibit the natural dynamics of the monitored processes. Therefore, targeting both nuclear and cytoplasmic properties of transcripts is troublesome unless either a single probe or multiple small probes are used. If a single non-transcript specific probe is used, issues may arise over how to differentiate the specific RNA the probe is binding.
Here we describe the application of the small fluorescent probe Pyronin Y (PY) to monitor transcript dynamics in living NIH3T3 cells. PY is a cationic environment-sensitive probe used to label endogenous double-stranded RNA (dsRNA) including mRNA, tRNA and ribosomal RNA (rRNA) (Cowden and Curtis 1983, Darzynkiewicz and Carter 1988, Darzynkiewicz et al 1986, Traganos et al 1988). The probe is sized below the exclusion mass of nuclear pore complexes (Grunwald and Singer 2010, Grunwald et al 2011) allowing entry into the nucleus.
PY labels all double-stranded RNA which enables an overall analysis of RNA functionality, structure, interaction and properties. This does, however, present concern over how to identify the different RNA subtypes labeled by the probe (how many different types of RNA subspecies are interacting with the probe and their position within living cells). Most analyses require a detailed understanding of probe/target interaction for each individual component the probe may bind to. Given the fragility of RNA species and the concerns over a loss of purity upon extraction (Bao et al 2009, Mitsuhashi et al 2006), we undertook a FLIM (fluorescence lifetime image microcopy) analysis using the phasor approach to determine the possible number of different RNA types interacting with this probe directly in live cells. The use of phasors has recently been applied to enhance the understanding of metabolic coenzymes (Stringari et al 2011, Wright et al 2012) as well as to examine the dynamics of fluorescent probes using the concept of spectral phasors (Fereidouni et al 2012).
In phasor FLIM, the decay curve of each pixel is measured and Fourier transformed to produce two coordinates for each pixel. These coordinates are plotted in the phasor plot. The phasor plot provides a graphical representation of the lifetime at each pixel without the requirement for fitting processes (Digman et al 2008). Within the plot, the universal circle provides a spatial reference for all attainable single exponential lifetimes. Pixels in the phasor plot could cluster. These clusters could represent well defined structures of the cell such as the nucleus or mitochondria or other cellular features. To assess if the clusters in the phasor plot correspond to specific structures of the cell, a region of the phasor plot is selected using a circular cursor of a given size and the corresponding pixels of the image are highlighted with the same color as the cursor. It is a simple task to see if the highlighted pixels correspond to a specific region of the cell, for example the nucleus, the nucleolus, the cytoplasm, the perinuclear region or specific puncti. The purpose of this study is to determine if we can recognize clusters of phasors which correspond to specific cellular substructures. This approach requires no prior knowledge of the decay of PY in different RNA transcripts.
2. Materials and methods
2.1. Cell culture
The NIH3T3 cell line was cultured in DMEM supplemented with PEN/STREP and 10% FBS. The cells were maintained in a water-jacketed incubator set to 37 °C and 5% CO2. MatTek dishes were coated with fibronectin (3 μg ml−1) 24 h prior to imaging and stored at 4 °C. The cells were plated on the dishes, exposed to PY (12 μM) and imaged.
2.2. RNA extraction
RNA was extracted from live NIH3T3 cells using the Ambion PureLink™ RNA Mini Kit as per the manufacturer's specifications and labeled with PY (24 μM). Quantitative analysis was carried out using a Nanodrop (Wilmington, US) with nucleic acid detection of 317 ng μl−1.
2.3. Microscopy
Each data set was acquired using the Olympus Fluoview 1000 laser scanning confocal microscope equipped with an ISS A320 FastFLIM (FLIMbox). For all measurements, an external Fianium laser was used with excitation of 545 nm (62 μW measured at the sample, collection bandwidth 560–660 nm). The pixel time was set to 20 μs with an image size 256 × 256. The Fianium laser repetition rate is 20 MHz.
A rhodamine 110 solution (2.1 μM) was used for system calibration with a known single exponential lifetime of 4.08 ns. Each FLIM data set was acquired with the aid of the SimFCS software (available from www.lfd.uci.edu/globals/ ). Typically about 107 counts were acquired over the entire image with integration of about 100 frames.
2.4. Phasor FLIM
The decay curve produced for each pixel is Fourier transformed to determine the two coordinates used to construct each phasor (Digman et al 2008, Stringari et al 2011, Van Munster and Gadella 2005, Wright et al 2012). The transformed data at a pixel level are then plotted onto the phasor plot where pixel clusters may be selected to highlight pixels in the image with similar fluorescence decays. The algorithms used to calculate the g (x-axis) and s (y axis) coordinates for the phasor plot are presented below, where i and j represent the position of the pixel, ω is the frequency where ω = 2πf, while f is the repetition rate of the laser (20 MHz).
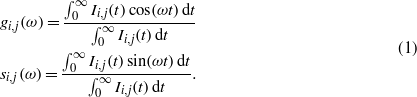
The g and s coordinates for single exponential lifetimes can be calculated through the use of equation (2) where τ is the lifetime of the component and ω is the laser frequency. Single exponential lifetimes always lie on the universal circle.

Multi-exponential lifetimes are characterized by expressions such as equation (3) where hk is the fractional intensity contribution of each single exponential component (τk) to the complex decay. When lifetime components come from multiple measured species (as is the case for multi-exponential lifetimes), the phasor of that species will lie inside the universal circle.

Manually inserted cursors of various sizes and colors were used to highlight different phasor clusters. The points selected in the phasor plot correspond to specific pixels in the image. In the images of the cells we create color-coded regions where each pixel in the image is colored according to a region of the phasor plot defined by the cursor center and the cursor radius. The center of the cursor in the phasor plot was used to calculate an average lifetime according to equation (4). Strictly speaking the position of the cursor can only be defined by both coordinates g and s. We define the average lifetime by the following formula

This formula calculates the point on the universal circle corresponding to the line from the origin passing through the center of the cursor intercepting the universal circle. We calculated for each cluster the average lifetime. Note that two clusters could appear very close from the average lifetime point of view but still occupy different positions in the phasor plot. Using this formula, we characterized the average lifetime of each cluster.
3. Results
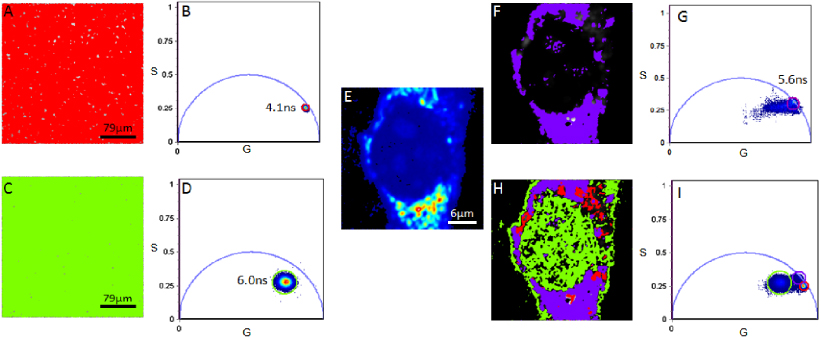
To assess if PY displays different decay properties when bound to dsRNA species in living cells, we first examined the binding of the probe in solution (figure 1(A)). Phasor plots were produced for PY in DMEM to establish the position of the phasor for PY not bound to a transcript. To measure fluorescence of the probe in DMEM, PY was used at a minimum of 5 × the concentration used for cell imaging, since the probe has very weak fluorescence when free. The determined lifetime of the solution was 4.1 ± 0.3 ns with the single cluster distribution always positioned on or just inward of the universal circle (figure 1(B)), indicating a single exponential decay for the probe non-interacting with RNAs. Extracted and purified RNA labeled with PY (figure 1(C)) displayed a lifetime of 6.0 ± 0.4 ns with a large cluster distribution positioned inward of the universal circle (figure 1(D)). The cells exhibited a longer lifetime when labeled with PY (figures 1(E)–(G)) with respect to PY in the DMEM. These measurements indicated that the probe has different decays when bound to a transcript (figures 1(H) and (I)).
Figure 1. The lifetime image and phasor distribution of PY in DMEM ((A) and (B)) and RNA extracted from living NIH3T3 cells labeled with PY ((C) and (D)). The average lifetime for PY in solution and extracted RNA in this instance were determined to be 4.1 ns (red cursor) and 6.0 ns (green cursor), respectively. Cluster selection of a PY labeled cell (E) is presented in (F) (lifetime image) and (G) (phasor plot). The contributions of all three labeled components (PY in solution: red cursor; extracted RNA labeled with PY: green cursor; a PY labeled cell: purple cursor) are presented in the phasor plot in (I) with the selection of each cluster of pixels within the plot corresponding to the color-coded lifetime image in (H). Note the linear combination of extracted RNA labeled with PY and a cell labeled with PY.
Download figure:
Standard imageWhen we go from the phasor plot to the image, like in figures 1(A) and (C), we use a '95% rule' in which we determine the radius of the cursor in figures 1(B) and (D) which maps to this percentage of pixels in the image. Then we apply a similar concept to complex images like that in figure 1(E). For example, let us assume that the image is divided into three regions of different fluorescence decay. For each region we could apply the same rule and use a given phasor cluster of a given radius and position to select 95% of the pixels of a given morphological region of the cell. Of course, this can only be done if the decay is sufficiently resolved in the three regions. Otherwise we might be able to identify only two regions as having different decays, or none. Using this method, for the complex phasor clusters in figures 1(G) and (E), we use the criteria to associate a selection of phasors to specific regions of the image. These regions are identified by morphological features such as the nucleus, the perinuclear region and the cytoplasm in figure 1(H).
In this example, we explore the phasor plot with our cursor and then look at the image. If there is a position of the cursor in the phasor plot that selects most of the pixels (95%) in the nucleus (in the image), then we associate this region of the phasor plot to the decay of the PY in the nucleus. The only reason why this separation could succeed is if there is a characteristic decay of PY in the nucleus that is different than the decay of PY in other parts of the image. Then it is reasonable to associate a particular region of the phasor plot (a cluster of phasors) to the nucleus. The same reasoning applies to the other identifiable structures in the cell. Each region is identified by a color. This color map is overlaid on the image (in grayscale) to associate clusters of the phasor with morphological features. Note that the green cluster in figure 1(I) is very large and it could contain identifiable sub-clusters as shown in the analysis of figure 2.
Figure 2. The intensity image (A) and lifetime images ((B)–(E)) with phasors chosen through cluster selection in the polar plot pictured in (G), ((H): zoomed) with reference to the intensity image. Four distinct phasor clusters were selected which included most labeled cell regions. The average lifetime for each cluster selection was: red cursor 5.74 ns, purple cursor 5.65 ns, green cursor 7.03 ns and cyan cursor 4.68 ns. The phasor selected colored images of all four manually selected regions is presented in (F). In the images with color-coded phasor selections the intensity image is given using a grayscale.
Download figure:
Standard imageThe examination of nucleic acid fluorescence decay labeled with PY in living cells occurred through manual selection of phasor clusters in each phasor plot, with reference to the intensity images of each cell, to establish a point of reference for each cell compartment. Within the polar plots, we identified at least four phasor clusters. What allows us to identify separate clusters in the overall broad distribution is the association of morphologically distinguishable regions of the cell with a specific part of the cluster. At times, these phasors were localized to a single region, for example the cytoplasm (figures 2(D) and 3(F)) while in other instances, the selection of a single phasor cluster led to the selection of pixels within different cell compartments. These compartments included both the nucleolus and peripheral regions in the cytoplasm (figures 2(B) and 3(C)) or the nucleus and some sporadic cytoplasmic regions (figures 2(C) and 3(D)).
Figure 3. Intensity image (A) and lifetime images ((B)–(F)) of RNA species in a single NIH3T3 cell labeled by PY. Cluster selection occurred through the phasor plot pictured in (H) ((I): zoomed) where color-coded cursors (yellow, red, green, purple and cyan) correspond to the images presented in (B)–(F). Five distinct phasor clusters were selected, outlining different regions of the cell (A). The average lifetime for each cluster selection was identified to be 4.77 ns (yellow cursor), 5.06 ns (red cursor), 6.13 ns (green cursor), 4.80 ns (purple cursor) and 3.68 ns (cyan cursor). The spatial distribution of the selected lifetimes is presented in (G). In the images with color-coded phasor selections the intensity image is given using a grayscale.
Download figure:
Standard imageThe observed dual localization patterns in the nucleus (green and red colors in figure 2(F)) were consistent for each nucleus in each living cell investigated (N = 30).
Further analysis of the main phasor cluster which maps to the cytoplasm leads to the extraction of additional cytoplasmic labeled species (figure 3). At least five different regions of the cell correspond to different clusters of phasors in the phasor plot. Some of the selections (cursor position in the phasor plot) highlight pixels in the nucleus but also in the cytoplasm. Other selections show clusters of pixels in the perinuclear region. It is notable that the FLIM signature of RNA transcripts is different in different regions of the cell.
Photostability of the fluorophore was observed, with changes in the focal plane not altering the detected lifetime of the species (figure 4). In this figure, three different focal planes were imaged at different times with about 1 min between images. The presence of most transcripts was observed to be constant as a function of time in the three images (figure 4) and the structures such as the nucleolus or mitochondria can be followed in the three planes.
Figure 4. The intensity images ((A), (C) and (E)) and lifetime images ((B), (D) and (F)) of transcripts in a single NIH3T3 cell labeled by PY. The three images (A), (C) and (E) were taken at different planes about 1 μm apart and at different times, about 1 min apart. Cluster selection occurred through the phasor plot pictured in (G) ((H): zoomed) where color-coded cursors show the selection of average lifetimes in the images. Photostability was observed overall with only small alterations in the detected lifetime of most cellular compartments as a function of time (A)–(D). Small changes in focal plane (about 3 μm) still showed consistency in lifetime detection, for example in the nucleolus (C)–(F). The detected average lifetime values for each cursor were 4.51 ns (red), 4.56 ns (green), 4.18 ns (purple), 4.89 ns (cyan) and 4.09 ns (yellow).
Download figure:
Standard image4. Discussion
Different RNA species in live cells may be assessed using the xanthene derivative probe PY to label all double-stranded RNA. The properties of PY labeling have been previously examined (Cowden and Curtis 1983, Darzynkiewicz and Carter 1988, Darzynkiewicz et al 1986, Traganos et al 1988) with a lifetime determination of 1.77 ± 0.01 ns for the probe in methanol (Toprak and Arik 2010). The lifetime of PY in living cells when bound to nucleic acid species has to our knowledge not been examined. Given that PY is sensitive to its environment, we propose that the lifetime properties of the probe may change with reference to the interacting transcript subtype. Thereby a lifetime analysis provides significant potential to enable differentiation of RNA transcripts.
We first identified the phasor cluster distribution position for PY in DMEM and PY labeled RNA previously extracted and purified from living NIH3T3 cells. We determined the average lifetime for each component to be 4.1 ± 0.3 ns and 6.0 ± 0.4 ns for PY in DMEM and extracted RNA respectively. From this we inferred that PY lifetime could alter when bound to a transcript. In agreement, PY labeled cells, where all double-stranded transcripts were targeted, led to an overall detected longer lifetime with a linear combination between a PY labeled cell and extracted RNA separately established (figure 1). There was only minimal detection of free PY within the live cells.
In the intensity images for all cells measured (N = 30), the labeled transcripts were positioned mainly within the cytoplasm and the nucleolus. Any further distinction of transcript subtype was not feasible through an intensity analysis and, therefore, we undertook a lifetime analysis in an attempt to extract further information on PY labeled RNA subtypes in live cell microenvironments.
Cluster differentiation in individual cells led to the identification of up to six PY labeled species with different detectable phasor locations. The detected phasor clusters appeared spatially separated and independent in the images. Lifetime clusters were either isolated within a single cell region such as the nucleus or cytoplasm or within multiple cell compartments. Transcripts detected in the nuclear regions alone exhibited the longest average lifetime of 6.06 ± 0.27 ns. This was the case for each analysis (N = 30 cells). The selection of lifetime clusters within the nucleolus of the living cells exhibited an average lifetime of 5.27 ± 0.23 ns. Selection of these clusters led to the selection of pixels not only in the nucleolus but also sporadically positioned in the cytoplasm. We propose that these may be a mixture of rRNA in the nucleolus and mRNP complexes in the cytoplasm due to the detected position of the species. Selected pixels were positioned both in the nuclear and cytoplasmic region with an average lifetime of 5.65 ± 0.57 ns. Pixels within this detected range were at times visualized to be within the nucleolus, suggesting possible labeling of rRNA, assembly of ribosomes within the nucleolus/nuclear regions and positioning of functioning ribosomes in the cytoplasm. Further, transcripts isolated in the cytoplasm alone had an average lifetime of 4.19 ± 0.65 ns.
Interestingly, the longest detected average lifetimes were located in small structures of a circular morphology seen in close proximity to the nucleolus. We propose these structures may be Cajal bodies due to their size, number and localization points (figures 4(D) and (F); cyan). The lifetime of these structures remained constant as a function of time. The species were seen to interact with the nucleolus (data not shown).
Overall, our results suggest PY labeled RNA species in live cells may be differentiated with the aid of phasors and FLIM analysis. We found that the phasor plot is consistent within each cell with the longest average lifetime detected in the nucleus of the live cells and the shortest in the cytoplasm. This is to our knowledge the first examination of RNA subtype classification using a single probe.
Acknowledgments
We thank Sohail Jahid for her assistance with RNA extraction and purification. This work is supported in part by NIH-P41-RR03155 and P41 GM103540 (EG, MD) and NIH P50-GM076516 (EG).