Abstract
A colorimetric sensing strategy employing gold nanoparticles and a paper assay platform has been developed for tuberculosis diagnosis. Unmodified gold nanoparticles and single-stranded detection oligonucleotides are used to achieve rapid diagnosis without complicated and time-consuming thiolated or other surface-modified probe preparation processes. To eliminate the use of sophisticated equipment for data analysis, the color variance for multiple detection results was simultaneously collected and concentrated on cellulose paper with the data readout transmitted for cloud computing via a smartphone. The results show that the 2.6 nM tuberculosis mycobacterium target sequences extracted from patients can easily be detected, and the turnaround time after the human DNA is extracted from clinical samples was approximately 1 h.
Export citation and abstract BibTeX RIS
Content from this work may be used under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
This paper describes a colorimetric sensing strategy employing unmodified gold nanoparticles (AuNPs) and microfluidic paper-based analytical devices (μPAD) for tuberculosis (TB) diagnosis. After mixing with the AuNPs colloid and then being triggered with sodium chloride solution, unmodified single-stranded deoxyribonucleic acid (ssDNA) detection sequences were directly hybridized with extracted double-stranded DNA (dsDNA) from TB patients or healthy persons without complicated AuNP probe preparation processes. The specific IS6110 dsDNA sequence of Mycobacterium tuberculosis complex (MTBC) was chosen as the diagnostic target for recognition [1]. When the target DNA sequences were absent, the detection ssDNA sequences were absorbed on the AuNP surfaces and protected unmodified AuNPs from aggregation in the high salt solution. The detection ssDNA sequences were hybridized with target DNA when the target DNA sequences were present, and the color of the unprotected AuNPs colloid turned from red to blue [2, 3].
Multiple detection results were simultaneously collected and concentrated from the paper assay platform. Data was readout by smartphone and analysis was performed via cloud computation to avoid the necessity for sophisticated detection equipment [4–6]. By using the proposed diagnostic platform, 2.6 nM MTBC target sequences could be detected via the color variance of AuNP spots on paper, and the turnaround time after human DNA is extracted from clinical samples could be as little as 1 h.
TB is a common and often lethal infectious disease caused by various strains of mycobacteria, usually M. tuberculosis in humans. Despite encouraging progress in reducing the mortality and incidence rates of TB, this disease remains a global public health problem. According to the World Health Organization (WHO), there were an estimated 8.7 million new cases of TB in the world, and approximately 1.4 million people died from TB in 2011 [7]. In addition, there has also been a resurgence of the disease worldwide because of the increase in immunocompromised states caused by AIDS, chemotherapy regimens or immunosuppressive therapy.
Skeletal tuberculosis may account for 35% of the extrapulmonary cases, and TB is often more likely to attack the spine. If not treated, spinal tuberculosis may cause spinal deformity or even paraplegia and pulmonary insufficiency. The clinical presentations of TB and bacterial spine infection share similarities, and both result in pain due to the loss of structural integrity. The infection may be confined to the central body (central pattern) and is often confused with malignancy. Diagnosing spinal TB is difficult because it is especially difficult to distinguish between TB infection, other bacterial infections and spinal tumors using x-ray, computed tomography (CT) and magnetic resonance imaging (MRI) assessments [8]. With spinal tuberculosis, therapy is highly dependent on early correct diagnosis, surgical intervention and a prolonged multidrug regimen.
Due to the majority of newly infected cases in developing countries where clinical laboratories and trained personnel for effective diagnosis and treatment are lacking, a rapid, simple, low cost and highly accurate on-site detection platform for spinal TB diagnosis in resource-constrained settings is needed to achieve early detection of tuberculosis for better patient management and infection control [9].
After more than a century of diagnostic method development since the identification of TB in 1882, a sensitive, rapid and cost-effective diagnostic platform still remains a challenge. Current TB detection methods mainly employ sputum smear microscopy, culture of bacilli and molecular species diagnostics [10–15]. In patients with TB, a biopsy specimen positive for smear microscopy in combination with Ziehl–Neelsen staining is a cost-effective way to obtain the results in minutes in high incidence areas. However, this identification method has a sensitivity of only approximately 50% (34–80%), especially in HIV-infected patients, and additional diagnostic methods need to be used to confirm negative results [10, 11]. Bacillus culture can be considered the standard method for TB detection in a clinical center. However, the result cannot be obtained instantly based on MTB because the bacillus requires 3–6 weeks for growth on solid culture media and 9–16 days using rapid liquid culture media [12, 13]. For this reason, polymerase chain reaction can be used to identify the mycobacterium with a sensitivity and accuracy of 95 and 92%, respectively. Molecular species diagnostics such as nucleic acid amplification tests adopt reverse-transcription polymerase chain reaction (RT-PCR) to amplify the specific MTB target DNA such as IS6110, 16S rDNA, cyp141, etc., from extracted sputum or tissue samples. These analyses possess the ability to achieve high accuracy, full automation and simultaneous detection of multiple MTB strains [14, 15]. However, test turnaround time including multiple hybridization processes requires 2–5 h, and the requirement of sophisticated infrastructure and trained personnel limits the feasibility of applying this test in the developing countries.
With the boost of various nanomaterial syntheses in combination with analytical platforms, AuNPs have been adopted as versatile biosensors to provide alternative schemes to conventional detection methods for TB diagnosis [16–19]. AuNPs and different shapes and composition of gold nanomaterials not only possess a high surface-to-volume ratio for binding high density biochemical molecules, they also have specific optophysical and various chemical characteristics for colorimetric sensing based on controlling the synthesized size, morphology and surrounding chemical environment [20–27]. The color of monodisperse AuNPs colloid (13 nm) is red because of the surface plasmon resonance (SPR), which is caused by the collective oscillation of the conduction electrons on the AuNP surfaces [28]. When the AuNPs aggregate, the decrease of interparticle distance between AuNPs induce surface plasmon coupling resulting in a red shift of the absorbance wavelength from 520 to 650 nm and a colloid color change from red to blue [29]. By manipulating electrostatic forces and steric repulsion between neighboring AuNPs, various colorimetric detection strategies have been developed for TB nucleic acid sensing. Soo et al adapted a DNA-mediated AuNP assembly initiated by Mirkin for identification of MTB and differentiation of MTB from other members of the MTBC from clinical sputum samples [16, 30]. Two designed ssDNA sequences that were complementary to both ends of the target DNA were modified separately on AuNPs. After hybridization of the target DNA with the probes, AuNPs probes were aggregated, and the color of the solution changed from red to purple. Salt-induced aggregation of AuNP probes for TB diagnosis was proposed by Baptista et al [17, 19]. AuNPs were also modified with thiolated detection ssDNA that was complementary to the target DNA. The presence of positive samples in the complementary AuNP probes prevented AuNPs aggregation in the high salt solution. In addition, a paper-based platform and smartphone were combined for on-site MTBC sensing [19]. However, the detection methods above used thiol-modified ssDNA sequences for the formation of AuNPs probes via Au–S bonding. Fabrication of AuNPs functionalized with thiolated DNA was not only complicated and took hours to complete, but also required well-trained personnel.
Here, we introduce a platform using unmodified AuNPs with an ssDNA detection probe and μPAD for rapid, cost-effective and high throughput TB diagnosis.
2. Experimental
2.1. Institutional review board approval
All tissue and human samples used in this report were approved and issued by the institutional review board (IRB) from Chang Gung Memorial Hospital.
2.2. Chemicals
Hydrogen tetrachloroaurate(III) trihydrate was purchased from Acros Organics (Geel, Belgium). Methanol, 2-propanol, sodium chloride, tris(hydroxymethyl)aminomethane, boric acid, ethylenediaminetetraacetic acid, ampicillin, deoxyribonucleotide triphosphate and trisodium citrate were ordered from Sigma-Aldrich (St Louis, MO). Oligonucleotides were synthesized and purified by Bio Basic Inc. (Bio Basic, Ontario, Canada). The water used in all experiments was ultrapure water purified in a Milli-Q system (Millipore, Milford, MA). Q solution, RNase, QIAamp DNA blood mini kits and HotStar Taq DNA Polymerase were purchased from Qiagen (Hilden, Germany). High Purity Viral Nucleic Acid Kit was obtained from Roche Applied Science (Mannheim, Germany). AxyPrep Maxi Plasmid Kit was purchased from Axygen Biosciences (Union City, CA). Novel Juice is ordered from Gene direx Inc. (Las Vegas City, NV). Whatman Grade 1 Chr cellulose chromatography paper was obtained from GE Healthcare (Little Chalfont, UK).
2.3. Instrumentation
The PCR amplification was performed in an Applied Biosystems 2720 Thermal Cycler (Life Technologies, CA, US). The size and morphology of synthesized AuNPs were verified by a transmission electron microscope (TEM; H7100, Hitachi High-Technologies, Tokyo, Japan). A UV–Vis spectrometer (Cintra 10e, GBC, Victoria, Australia) was used to obtain the extinction spectra of the AuNPs colloid for concentration and dispersion/aggregation examination. The PCR amplification was performed in Applied Biosystems 2720 Thermal Cycler (Life Technologies, Carlsbad, CA, USA). The AuNPs spots of TB diagnostic results on chromatography paper were recorded using a smartphone (butterfly, HTC, Taiwan) under artificial white light without flash. All the images were then uploaded to a cloud storage (Dropbox, San Francisco, CA) via internet followed by red, green and blue (RGB) value analyses using free software ImageJ (National Institutes of Health, US).
2.4. Preparation of AuNPs
The citrate-capped AuNPs were synthesized according to the citrate reduction of HAuCl4 [31]. To achieve the reduction, a 25 ml solution containing 38.8 mM trisodium citrate was added to 250 ml of a boiled 1 mM HAuCl4 solution. Following continuous heating for an additional 15 min, the AuNPs colloid was prepared when the color of the solution changed from yellow to a deep red. The diameter of synthesized AuNPs was 13 ± 1 nm, as confirmed by TEM images. The prepared concentration of AuNPs colloid for TB diagnosis was estimated by analyzing the extinction spectra using the Beer–Lambert Law [32].
2.5. Health control DNA extraction
The whole blood samples were collected from healthy controls and placed into tubes with EDTA buffer. Genomic DNA was extracted using QIAamp DNA blood mini kits (Qiagen, Valencia, USA) according to the manufacturer's instructions. The extracted DNA was dissolved in tris-borate buffer (pH 9.0) to a final concentration of 15 ng μL−1 and stored at −20 °C until further use.
2.6. Construction of 123 bp amplicon of IS6110 target dsDNA sequence
Luria Bertani (LB) plates containing the standard plasmid and the 123 bp fragment were obtained from Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou. The white colonies were selected, then grown in LB broth containing 50 μg ml−1 Ampicilin and incubated in 37 °C shaker at 225 rpm overnight. DNA was isolated from the culture using the AxyPrep Maxi Plasmid Kits and amplified using HotStar Taq DNA Polymerase to confirm the presence of the 123 bp fragment. Ten micro liters of DNA template was used in a 50 μl reaction mixture containing 0.6 μl of each 25 μM primer, 4 μl of 2.5 mM dNTP, 5 μl of 5 × Q solution, 5 μl of 10 × buffer, 0.25 μl of 5U μl−1 HotStar Taq and 24.55 μl of RNase free H2O. The temperature cycling conditions for PCR amplification were as follows: initial denaturation at 94 °C for 15 min, 35 cycles of 94 °C for 1 min, 68 °C for 1 min, 72 °C for 1 min, and a final extension of 72 °C for 10 min. The amplified products were analyzed by gel electrophoresis in 2% agarose prepared in TBE buffer, added Novel Juice to loading samples, and visualized on a UV-light transilluminator.
2.7. Paper-based colorimetric diagnosis of TB
The TB diagnosis was performed using salt-induced colorimetric development of the AuNPs colloid with analyte solutions followed by data readout using a paper-based platform and a smartphone. The hydrophobic barrier was formed by printing solid wax on the chromatography paper and then heating to 140 °C for 2 min on a hot plate. Detection oligonucleotide sequences were dissolved (100 mM) in tris-borate buffer (50 μl, pH 9.0) to form a 1.3 nM probe solution. IS6110 (2.6 nM, 0.52 pmol) and healthy control DNA sequences (2.6 nM, 0.52 pmol) were prepared in tris-borate buffer (50 μl, pH 9.0) and were then added to detection oligonucleotide solutions, followed by a hybridization process that included denaturalization at 90 °C for 1 min and annealing at room temperature for 4 min. A 1 M sodium chloride solution (50 μl) was added to the analyte mixture, and 850 μl of 0.5 nM AuNPs colloid was then added. After 30 min of color development, the mixture was then pipetted onto the chromatography paper with a solid wax forming the hydrophobic barriers and color standard for image analysis.
3. Results and discussion
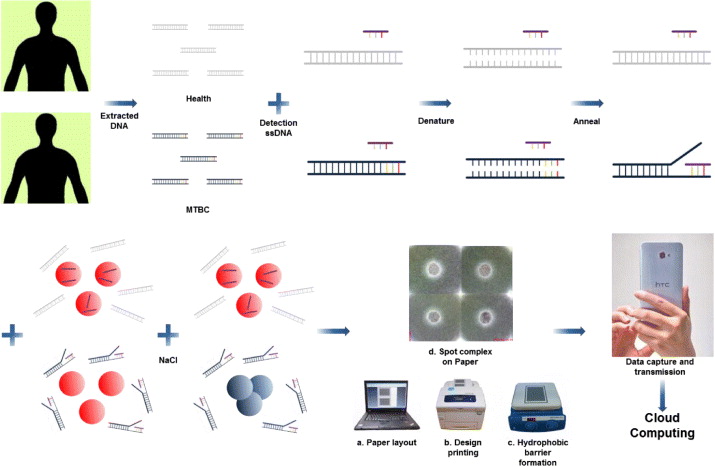
Based on the SPR effect, the color of the gold colloid may be correlated to the degree of aggregation of AuNPs in aqueous solution [33]. The status of the aggregation depends on the surface charge and steric effect on the AuNPs surfaces that can be manipulated by introducing salt into the solution or binding biomolecules via electrostatic adsorption, high affinity Au–S or Au–N bonding or control of the medium properties and composition [34, 35]. In this report, a salt-induced colorimetric sensing strategy employing unmodified AuNPs and a paper assay platform for TB diagnosis was adopted. The schematic illustration of the TB diagnosis combining salt- induced AuNPs sensing and the μPAD is shown in figure 1. The detection oligonucleotide sequences were first hybridized with extracted DNA sequences from patients or healthy controls from healthy humans, followed by addition to the AuNPs colloid and induction of colorimetric sensing with a sodium chloride solution. Unmodified ssDNA sequences were used to replace the costly thiolated ssDNA as detection probes, and the complicated preparation procedure for AuNPs conjugated molecular probes was eliminated to shorten the diagnosis time. The mixture was then spotted and concentrated on the chromatography paper confined by solid wax hydrophobic barriers. A smartphone was used to take images of the diagnostic results and transmit these images to a server for cloud computing.
Figure 1. The schematic illustration of the proposed TB diagnostic. Unknown extracted human DNA sequences were first hybridized with detection oligonucleotide sequences followed by addition of AuNPs colloid and triggering of the colorimetric sensing with a sodium chloride solution. If the extracted DNA sequences consisted of IS6110 target sequences, the detection oligonucleotide sequences would hybridize with them, and only a few ssDNA sequences would be absorbed on the AuNP surface to avoid aggregation after the addition of salt. In the absence of IS6110 target sequences, the color of the mixture remains red after hybridization and does not change. The mixture was then spotted, and concentrated on the chromatography paper confined by solid wax hydrophobic barriers. A smartphone was used to take images of the diagnostic results and transmit to a server for cloud computing.
Download figure:
Standard image High-resolution image3.1. Colorimetric sensing of TB using unmodified AuNPs and ssDNA for TB diagnosis
The AuNPs are dispersed in an aqueous solution based on the interparticle repulsion induced by the negative surface charge of AuNPs synthesized using the citrate reduction method [36]. When a high salt solution is added, the dissociative ions shelter the electrostatic repulsion and result in the aggregation of AuNPs [37]. However, if detection ssDNA sequences are added to the AuNP colloid before the addition of salt, the uncoiled ssDNA would absorb on the AuNP surface based on the hydrophobic interaction [38], exposing the negatively charged phosphate backbone to avoid aggregation. In contrast, dsDNA added to the AuNP colloid would not absorb on the negatively charged AuNPs surfaces because dsDNA possesses a stable double-helix structure that presents a negatively charged phosphate backbone, so dsDNA would not prevent the aggregation of AuNPs in a high salt solution [2, 3]. The different optical properties of the salt-induced AuNPs aggregation can be adopted for DNA-hybridization-based biomolecular recognition assays.
In this report, unmodified 13 nm AuNPs were used, and a 24-mer oligonucleotide sequence, 5'-CTC-CGT-CCA-GCG-CCG-C-3', which is complementary to the IS6110 target DNA sequence, was chosen as the detection ssDNA sequence. The presence of the123 bp fragment indicates the positive test for the M. tuberculosis complex, so IS6110 dsDNA was served as the positive target sequence to verify the proposed colorimetric assay in this study. When the extracted DNA samples consist of the IS6110 target sequence, detection ssDNA would hybridize with the target DNA sequence and would not protect the AuNPs from aggregation after adding a high salt solution. In contrast, the detection ssDNA sequence would suspend and attach on the AuNP surface to serve as protection if the test DNA was extracted from a healthy human.
A moderate amount of 25 mM salt was chosen to be used in the following tests according to the colorimetric sensing mechanism. The amount was chosen between a large amount of salt (> 0.2 M) causing AuNP aggregation even with ssDNA protected and an insufficient amount of salt (< 10 mM) introducing no color variance of the unmodified AuNPs colloid. The degree of aggregation of the AuNP mixture was first measured via UV–Vis spectroscopy to verify the proposed colorimetric sensing mechanism (figure 2). The absorbance wavelength of the unmodified and monodisperse AuNP colloid was centered at approximately 520 nm. The presence of 25 mM salt induced the aggregation of the AuNPs, and the absorbance wavelength of the AuNP colloid was red-shifted to ∼ 610 nm. In the absence of IS6110 target sequences, ssDNA detection sequences do not hybridize with healthy human DNA, and they do adsorb on the AuNPs surface to prevent the aggregation of AuNPs induced by the addition of salt. If IS6110 target DNA sequences are present, ssDNA would be hybridized with them, and aggregation of AuNPs induced by adding salt would result in an increase in the 520/610 nm extinction ratio to 2.4 compared to 1.7 in the absence of the IS6110 target sequence.
Figure 2. UV–Vis extinction spectra of unmodified AuNPs-based TB sensing. (I) The absorbance wavelength of the unmodified 13 nm AuNPs colloid centered at approximately 520 nm. (II) After addition of 25 mM salt into the AuNPs colloid, the salt induced the aggregation and the absorbance wavelength of the AuNPs colloid was red-shifted to ∼ 610 nm. (III) In the presence of 2.6 nM IS6110 target sequences, most of the detection oligonucleotide sequences would hybridize with them and would not provide extra negative charges on the AuNP surfaces to avoid aggregation when 25 mM salt was added. (IV) In contrast, the healthy control does not contain IS6110 target sequences, so the 1.3 nM detection ssDNA sequences would absorb on the AuNP surface to resist aggregation when 25 mM salt is added. The extinction ratio of Ext520/610 increases to ∼ 2.4 comparing to 1.7 for case (III). The inserted TEM image shows the diameter of synthesized AuNPs is 13 ± 1 nm.
Download figure:
Standard image High-resolution image3.2. Paper assay platform with unmodified AuNPs colorimetric sensing for TB detection
To achieve on-site TB diagnosis without the use of sophisticated equipment, a paper assay platform was adopted for the detection and readout of results (figure 3). A paper assay platform has several advantages such as low cost, ease of fabrication, compatibility with biological samples and compatibility with a wide variety of functionalities, which make it suitable for resource-constrained settings [4–6]. The color of the paper is white, so paper is a perfect candidate for result readout for colorimetric sensing. Solid wax was printed on the paper and heated to form the hydrophobic barriers through the paper cross section so that the AuNP mixture would be concentrated in the confined circle area and would reveal the colorimetric results. The amount of the test mixture used to spot on each paper assay circle was 200 μl, an amount that is smaller than the typical test mixture amount for UV–Vis spectra, which requires at least 1 ml of test mixture.
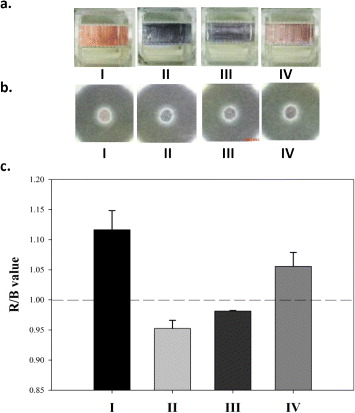
Figure 3. The colorimetric results of TB diagnosis (a) in cuvettes, (b) on paper and (c) the analytical values of red/blue of the spots. The four different solutions are (I) unmodified AuNPs colloid, (II) AuNPs colloid with 25 mM salt, (III) in the presence and (IV) absence of 2.6 nM IS6110 target sequences hybridized with detection ssDNA sequences in AuNPs colloid and then adding 25 mM salt. The colors in the cuvette are (I) red, (II) blue, (III) blue and (IV) red, and the mean R/B values of the spots on the paper are approximately 1.12, 0.96, 0.98 and 1.06, respectively (n = 3). The test conditions are identical to the UV–Vis extinction spectra test of figure 2 except the volume needed for UV–Vis was 1 ml and the spotting volume in paper was 200 μl.
Download figure:
Standard image High-resolution imageAnalyzing the AuNP mixture spotted on the paper using a smartphone and cloud computation, the red-green-blue (RGB) value red/blue of the spots in the presence of IS6110 target sequences was < 1, and the red/blue value of the spots was > 1 when the test DNA lacks the IS6110 target sequence. Compared to traditional clinical molecular species diagnostics that require several hours to finish the tests, the turnaround time for the proposed platform can be effectively completed in 1 h after DNA extraction. Thousands of test results can be analyzed via cloud computing and entered into the database of the clinical center for disease management and long-term post-therapy monitoring.
4. Conclusions
TB diagnosis remains challenging because the diagnosis needs to be sensitive, specific, and (especially) cost-effective based on the long-term post-treatment monitoring and the vast numbers of infected individuals in developing countries. In this report, we utilized salt-induced AuNP colorimetric diagnosis for sensing target TB DNA sequences without the need for multiple PCR cycles to amplify specific MTB target DNA sequences from extracted sputum or tissue samples. From our results, it is clear that unmodified AuNPs can be used to avoid complicated and time-consuming AuNP probe preparation processes, and costly thiolated ssDNA can be replaced by unmodified ssDNA. In addition, μPAD can be used for simultaneous screening of multiple results and analysis to achieve high throughput and to eliminate the requirement for sophisticated optical detection equipment and well-trained technicians. We found that, 2.6 nM MTBC target sequences could be detected, and the diagnosis after DNA extraction could be completed in 1 h. The ability to use a smaller amount of sample for testing and a lower concentration of TB diagnostics can be achieved by using AuNPs in a catalytic reaction to amplify the colorimetric signal and more compact paper assay areas. In addition to using IS6110 for MTBC diagnosis, different series of target DNA sequences can be adopted to eliminate the cross interference with other members of the mycobacterium genus and to further reduce the possibility of false positive or false negative results. We are still investigating the optimum parameters for the platform and performing blind tests using extracted DNA from clinical samples compared to the bacillus culture results. We believe the proposed platform possesses the potential for affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and highly end-user-deliverable (ASSURED) diagnostic applications.
Acknowledgments
This work was supported by Chang Gung Memorial Hospital, Linkou under Grant number CMRPG3G0821 and National Science Council, Taiwan (NSC 101–2113-M-005–001-MY2). We thank Pi-Yeih Chang and Mei-Chia Wang from Department of Laboratory Medicine of Chang Gung Memorial Hospital, Linkou for providing the standard strain of M. tuberculosis and technical support. We also thank Hung-Chen Fang and Natalie Yi-Ju Ho for their assistance in performing the experiments.