Abstract
The endeavor of exploiting arrays of vertical one-dimensional (1D) nanostructures (NSs) for cellular applications has recently been experiencing a pronounced surge of activity. The interest is rooted in the intrinsic properties of high-aspect-ratio NSs. With a height comparable to a mammalian cell, and a diameter 100–1000 times smaller, NSs should intuitively reach far into a cell and, due to their small diameter, do so without compromising cell health. Single NSs would thus be expedient for measuring and modifying cell response. Further organization of these structures into arrays can provide up-scaled and detailed spatiotemporal information on cell activity, an achievement that would entail a massive leap forward in disease understanding and drug discovery. Numerous proofs-of-principle published recently have expanded the large toolbox that is currently being established in this rapidly advancing field of research. Encouragingly, despite the diversity of NS platforms and experimental conditions used thus far, general trends and conclusions from combining cells with NSs are beginning to crystallize. This review covers the broad spectrum of NS materials and dimensions used; the observed cellular responses with specific focus on adhesion, morphology, viability, proliferation, and migration; compares the different approaches used in the field to provide NSs with the often crucial cytosolic access; covers the progress toward biological applications; and finally, envisions the future of this technology. By maintaining the impressive rate and quality of recent progress, it is conceivable that the use of vertical 1D NSs may soon be established as a superior choice over other current techniques, with all the further benefits that may entail.
Export citation and abstract BibTeX RIS

Content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
Understanding cell function, analyzing cell content, and investigating the intricate cell signaling pathways are of great interest for both fundamental studies and the continued understanding of pathologies. In addition, the possibility to modify signaling pathways and to record the subsequent cellular response is imperative for drug discovery. However, due to cellular complexity, poor delivery efficiency to many cell types, and current probes being on the same size scale as the cells they aim to investigate, gaining access to detailed cellular information remains a challenge to this day. More than a decade ago, it was postulated that by merging biology and nanotechnology, many of the limitations of current methodologies could be circumvented.
1.1. Combining nanostructures with mammalian cells
High-aspect-ratio objects with nanoscale diameters and microscale heights are generally referred to as one-dimensional (1D) nanostructures (NSs). They were originally developed for applications in physics to study quantum phenomena and single electron transport and were later also used for solar cells and capacitors [1–3]. However, the potential of using vertical 1D NSs for biological applications has been, and still is, an area gaining increasing attention and interest. The major motivation behind this development is a combination of a size compatibility of cells and NSs, along with the promising scientific, social, and economic possibilities any successful applications could entail. The compatibility of cells and NSs is rooted in the fact that 1D NSs are comparable in length to the size of a cell, and about 100–1000 times smaller in diameter than a cell. Thus, a vertical NS is an object potentially long enough to reach deep into a cell and thin enough to do so without causing damage. Many interesting cellular applications can be envisioned from combining cells and NSs. NSs could deliver [4–6] or collect material from different subcellular compartments and investigate cells otherwise hard to access [7]. The NSs could measure cell composition and activity, either directly owing to their electrical properties [8, 9], through delivery of molecular sensors [10], or by physically probing cellular components [11, 12]. The NSs could also modify the cell activity directly [8, 9], or by delivering a variety of molecules [4–6]. In addition, NSs could be used to modulate [13–15] and even measure [16–18] the adhesion of cells on surfaces.
By up-scaling from a single NS to groups of NSs in random or ordered arrays, one can envision powerful platforms for life science applications. NSs in an array format can allow many cells to be addressed in parallel [8, 9], which can be useful for investigating cell networks or for cell diagnostics. Arrays of NSs can also allow individual cells to be contacted by many NSs simultaneously [8, 9], thereby potentially providing spatial resolution across the cell body. If these multiple contact points across a cell network or cell body also allow interrogation over time [9], the NS array platform would provide temporal resolution in a vast number of cells simultaneously. This would entail gaining unprecedented information on detailed cell activity, which would be highly valuable for the basic understanding of cell signaling and drug discovery. The positioning of the NSs within the array could also be designed to create cellular networks by guiding cells and their processes along the NS pattern [19–21].
Rooted in the very first papers addressing this research subject a decade ago [22, 23], this review is a summary of the intense progress of interfacing cells with NSs that the last few years have brought about. Although both suspensions of NSs [24] and single NSs in field-effect-transistor devices [25] have many interesting biomedical applications, this review will cover arrays of vertical and semivertical high-aspect-ratio NSs for cellular applications. The term 'array' is used in a broad sense to cover groups of NSs with different regularity, i.e., both randomly scattered and orderly arranged NSs. Short NSs with heights below 500 nm or larger structures with widths above 500 nm fall outside the scope of this review. This review covers the broad spectrum of NS materials and dimensions used; the observed cellular responses with specific focus on adhesion, morphology, viability, proliferation, and migration; compares the different approaches used in the field to provide NSs with the often crucial cytosolic access; covers the progress toward biological applications; and finally, the future of this technology is envisioned.
2. Spectrum of NS arrays interfaced with cells
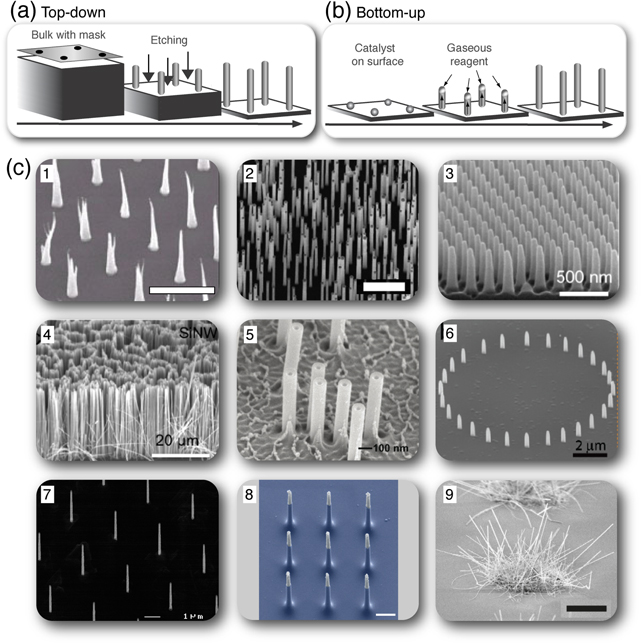
One-dimensional NSs can be fabricated top-down or bottom-up using a variety of methods [26]. Top-down methodologies entail etching NSs from bulk material, [figure 1(a)] and bottom-up techniques rely on the growth of vertical NSs from catalytic particles [figure 1(b)]. With either technique, it is possible to produce NSs of different materials and geometries (shape, diameter, height) in arrays with different density (number of NSs per area) and regularity (random or ordered arrays) (figure 1(c), table 1).
Figure 1. Methods for NS fabrication and a range of NS arrays used for cellular applications. NSs can be fabricated in arrays of either random or ordered NSs. The most common ways to make NS arrays are either to use top-down fabrication (a), where a mask is used and NSs are etched out from a bulk, or to use bottom-up fabrication (b), where NSs are grown from nucleation sites on a surface that collects the vapor phase atoms. (c) Scanning electron microscopy (SEM) images showing the diversity of NS arrays developed for cellular applications: (c1) ordered array of carbon NSs [Reproduced with permission from [22]. Copyright IOP Publishing.], (c2) random array of gallium phosphide NSs [Reproduced with permission from [27]. Copyright Elsevier 2013.], (c3) ordered array of quartz NSs [Reproduced with permission from [28]. Copyright IOP Publishing.], (c4) ordered array of silicon NSs [Reproduced with permission from [28]. Copyright IOP Publishing.], (c5) random array of hollow aluminum oxide NSs [Adapted with permission from [6]. Copyright 2012 American Chemical Society.], (c6) platinum NSs positioned in a ring [Adapted with permission from [29]. Copyright 2010 American Chemical Society.], (c7) ordered array of indium arsenide NSs (personal image), (c8) silicon NSs positioned in a 3 × 3 pad, reprinted with permission from Macmillan Publishers Ltd: Nature Nanotechnology [8], copyright 2012, and (c9) patches of semivertical silicon NSs, reprinted with permission from Macmillan Publishers Ltd: Scientific Reports [16], copyright 2013. Scale bar in (c1) is 5 μm, in (c2) is 1 μm, in (c8) is 1 μm, and in (c9) is 5 μm.
Download figure:
Standard image High-resolution imageTable 1. A summary of the NS arrays for cell interfacing purposes found in the literature. The NS core material, approximate shape, geometry, topography, and the type of cells, with which the NS arrays are interfaced, are given. The NS density is defined as low (density ≦̸ 1 NSs/100 μm2, spacing ⩾ 10 μm), medium (1 NSs/100 μm2 < density < 30 NSs/100 μm2, 2 μm < spacing < 10 μm), and high (density ⩾ 30 NSs/100 μm2, spacing ≦̸ 2 μm), where 'ordered' indicates an ordered array of NSs, 'random' indicates a random array of NSs, N.A. = not applicable (e.g., the NSs are positioned along a single line or in a pad smaller than a single cell). Material abbreviations: PS = polystyrene, PLGA = poly-(lactic-co-glycolic-acid). Cell abbreviations: CHO = Chinese hamster ovary cells, PC-12 = rat pheochromocytoma cells, HEK293 = human embryonic kidney cells, HeLa = human cervical cancer cells, PC-3: human prostate cancer cells. Numbers in square brackets correspond to numbers in reference list and * = semivertical NSs.
| Geometry | ||||||
|---|---|---|---|---|---|---|
| Topography | ||||||
| Core | ~Shape | Height (μm) | Diameter (nm) | Regularity, density | Cell type | Reference |
| C | Cone | 6–10 | 20–50 (tip) | Ordered, medium | CHO cells | [22] |
| Not provided | <100, 200 (tip) | Ordered, medium | Mouse myeloma cells, CHO cells | [23] | ||
| 7–10 | 100 (tip) | Ordered, N.A. | Rat hippocampal cells, PC-12 cells | [34] | ||
| 7 | <100 (tip) | Ordered, medium | None | [31] | ||
| 10 | 100 (tip) | Ordered, N.A. | Rat hippocampal tissue slice | [35] | ||
| 10–17 | 100 (tip) | Ordered, low-medium | CHO cells | [30] | ||
| 7–10 | Not provided | Ordered, not provided | Chinese hamster lung fibroblast cells | [32] | ||
| 15 | 250 (tip) | Ordered, medium | Chinese hamster lung fibroblast cells | [33] | ||
| 10 | Not provided | Ordered, N.A. | E.g. primary rat neurons, rat hippocampal tissue slice | [36] | ||
| Si | Needle | 7–12 | 100–300 (tip) | Random, medium-high | Rat and human hepatoma cells | [37] |
| Cylinder | 3–6 | 30, 90, 400 | Random, medium | Mouse embryonic stem cells, HEK293 cells | [43] | |
| >10 | 160 | Random, not provided | Mouse mesenchymal stem cells | [39]* | ||
| 20 | 100 | Random, medium-high | Human hepatic cells and hepatoma cells | [46]* | ||
| 2, 2.5, 3 | 140, 280 | Ordered, not provided | E.g. primary neonatal rat mechanocytes, HeLa cells | [18] | ||
| 10 | 100–200 | Random, high | Mouse immune cells (reacted with antibodies) | [41]* | ||
| Not provided | Not provided | Ordered/random, not provided | E.g. primary rat neurons and primary human fibroblasts | [5] | ||
| 2.5 | 20–80 | Random, not provided | CHO cells | [45] | ||
| 3 | 150 | Ordered, high | Primary rat neurons, HEK293 cells | [8] | ||
| 1–>3 | <150 | Ordered/random, medium-high | Primary mouse and human immune cells | [7] | ||
| 9, 14, 20, 26 | 162, 171, 175, 192 | Random, not provided | Primary human mesenchymal stem cells | [44] | ||
| 0.5, 25–30 | 140, 10–100 | Ordered/random, not provided | Human lung carcinoma cells | [28] | ||
| 25–30 | 10–100 | Random, not provided | Human lung carcinoma cells | [40] | ||
| 1 | 70–100 | Ordered, medium | Primary rat embryonic neurons | [20] | ||
| 5, 10 | 200, 400 | Ordered, medium-high | E.g. primary human mesenchymal stem cells, PC-12 | [14] | ||
| 3–5 | 100 | Random, medium | HeLa cells, PC-3 cells | [10] | ||
| 4.8 | 210–530 | Random, low-medium | Algal cells | [47] | ||
| 5–10 | 40 | Random, N.A. | Primary human fibroblasts | [16]* | ||
| >10 | 166, 85 | Random, not provided | Mouse mesenchymal stem cells | [38]* | ||
| 1.3 | 270 | Random, high | HeLa cells | [42] | ||
| 0.99, 1.17 | 80, 70 | Random, high | Mouse embryonic fibroblasts | [48] | ||
| 5 | 20–100 | Random, not provided | Human lung carcinoma cells | [49] | ||
| 9.5, 15.5 | Not provided | Random, not provided | Mouse mesenchymal stem cells | [51] | ||
| 1 | 150 | Random, not provided | HeLa cells, HEK293 cells, mouse embryonic fibroblasts | [52] | ||
| 9.5 | 83 | Random, low-high | Mouse mesenchymal stem cells | [50] | ||
| Pt | Cylinder | 0.7–2 | 75–400 | Ordered, medium/N.A. | Primary rat embryonic neurons | [29] |
| 1–2 | 150–200 | Ordered, N.A. | Mouse cardiac muscle cells | [9] | ||
| GaP | Cylinder | 2.5 | 50 | Random, high | Primary mouse neurons | [54] |
| 1.5–3 | 70 | Ordered, N.A. | Primary mouse neurons | [21] | ||
| 4 | 75 | Ordered, N.A. | Primary mouse neurons | [19] | ||
| 2.5–5 | 40, 80 | Ordered, high | Primary mouse neurons | [17] | ||
| 0.5, 1, 4 | 20, 40, 80 | Random, medium-high | Primary mouse retinal cells | [27] | ||
| 1.5, 3.8, 6.7 | 80 | Random, high | Mouse fibroblasts | [55] | ||
| GaN | Cylinder | Not provided | Not provided | Random, not provided | PC-12 cells | [56] |
| InAs | Cylinder | 1–3 | 100–300 | Random, medium | HEK293 cells, rat embryonic neurons | [53] |
| 2, 6, 11 | 100 | Ordered, medium | HEK293 cells | [57] | ||
| 4.4 | 92 | Ordered, low-high | HEK293 cells | [13] | ||
| IrOx | Tube | 0.5 | 181 (outer) | Ordered, not provided | Mouse and rat cardiac muscle cells | [70] |
| SiO2 | Cylinder | 0.5–2 | 50–500 | Ordered, medium-high | Primary rat embryonic neurons | [63] |
| 0.5 | 140 | Ordered, not provided | Human lung carcinoma cells | [28] | ||
| Tube | 5 | 500 (outer) | Ordered, medium | HEK293 cells, mouse embryonic fibroblasts | [4] | |
| ZnO | Cylinder | 0.5 | 50 | Random, high | E.g. mouse embryonic fibroblasts, bovine endothelial cells | [61]* |
| 0.5 | 40–50 | Random, high | Mouse embryonic fibroblasts | [84]* | ||
| 0.5 | 50 | Random, not provided | Primary mouse macrophages | [62]* | ||
| 2 | 200 | Random, not provided | PC-12 cells, rat embryonic myoblasts | [60]* | ||
| CuO | Cylinder | 0.5–5 | 100–150 | Random, not provided | HEK293 cells, HeLa cells | [59]* |
| Al2O3 | Tube | 1–2 | 100, 250 (outer) | Random, low-high | HeLa cells, CHO cells | [6] |
| 2.5–4 | 180 (outer) | Random, not provided | Mouse fibroblasts | [66] | ||
| 1.5 | 250 (outer) | Random, medium | HEK293 cells, CHO cells | [67] | ||
| 1 | 100 | Random, high | CHO cells | [68] | ||
| GaP/GaInP | Cylinder | 2.5–4 | 40, 80 | Random, not provided | Mouse fibroblasts | [58] |
| PS | Cylinder | 0.5 | 200 | Ordered, high | Mouse osteoblast precursor cells | [64] |
| PLGA | Cylinder | 0.8, 0.84 | 90, 250, 500 | Ordered, high | Primary human blood platelets | [65] |
The resulting vertical or semivertical NSs are classified as nanofibers, -rods, -tubes, -pillars, -wires, and -needles, but the diversity implied by these terms does not necessarily reflect the actual diversity of the NSs used for cellular applications. Whereas the terms 'fiber,' 'needle,' or 'tube' provide information on the NS shape, the essential difference between 'nanowire,' '-rod,' and '-pillar', which are all used to describe solid and cylindrical NSs, is less obvious. 'Pillar' and 'rod' are mostly used with lower-aspect-ratio NSs, but not consistently. Therefore, the NSs will simply be distinguished by approximate shapes in this review.
A wealth of materials has been explored for the fabrication of NSs for cellular applications (table 1). These materials may be conducting, semiconducting, insulating, or fluorescent and each has different advantages and drawbacks in terms of biocompatibility and controllability of NS growth. Thus, care should be taken in choosing the appropriate material for a specific application.
The first NSs grown for cell interfacing purposes were conical carbon fibers with tips as thin as 20 nm, but a micron-sized base, and heights of several microns [22]. These NSs, which are grown with the aid of catalytic nickel particles, have been precisely positioned to obtain simple square arrays [22, 23, 30–33] or single long rows of NSs [34–36]. Today, however, silicon is the most frequently used material. It has been shaped into pointy nanoneedles [37] and near-cylindrical NSs of higher or lower aspect ratio [5, 7, 8, 10, 14, 16, 18, 20, 28, 38–52], mostly through top-down fabrication; however, in a few cases, it was formed by bottom-up growth [16, 38, 39, 41, 43]. Silicon NSs may be positioned in ordered arrays [5, 7, 8, 14, 18, 20, 28], but are most commonly fabricated in random arrays with densities from 1–1000 NSs/100 μm2 [5, 7, 10, 28, 37, 40, 42–52], covering the entire range of NS densities (NSs/100 μm2) used for cellular applications, from low (≦̸1), over medium (>1<30), to high (⩾30). Furthermore, some silicon NSs have been grown as dense patches [16] or grass-like carpets of semivertical rods [38, 39, 41].
NSs made of other semiconducting materials, such as gallium phosphide, gallium nitride, and indium arsenide, with typical dimensions of around 100 nm in diameter and a few to several microns in height, may also be grown in random [27, 53–56] or ordered [13, 17, 19, 21, 57] patterns via random or highly controlled positioning of catalytic particles, respectively. Furthermore, growth of fluorescent gallium phosphide/gallium indium phosphide heterostructural NSs has been recently demonstrated [58].
Advanced patterns have also been created with lower-aspect-ratio platinum NSs, which have been positioned with high precision in a ring [29] or as ordered patches of a few NSs [9, 29]. Other structures include random arrays of copper oxide NSs of varying height [59], dense random arrays of semivertical zinc oxide NSs [60–62], and ordered arrays of lower-aspect-ratio quartz [28, 63], polystyrene [64], and poly-(lactic-co-glycolic-acid) [65] NSs.
Whereas all of the preceding structures are solid, hollow NSs have also been fabricated from silica [4], aluminum oxide [6, 66–68], aluminum indium phosphate [69], or iridium oxide [70] by deposition of material within a porous membrane or onto NSs followed by etching of the membrane or core structure, respectively.
The core material of the NSs and the associated methods of fabrication ultimately define the overall geometrical limits of the arrays, but many postfabrication modifications can be made to tailor the NSs to the given application or increase their biocompatibility. For instance, silicon NSs may have a calcium phosphate coating [38, 39] or may be silanized and coupled to a variety of molecules, ranging from smaller molecules to macromolecules, such as antibodies and DNA plasmids [5, 7, 10, 28, 40–42, 52]. Cytotoxic copper oxide can be encapsulated in a harmless silicon oxide coating [59], and cell attachment to the various materials can be improved by oxygen plasma treatment to increase the wettability of the surface [64] or by adsorption of adhesion-promoting molecules, such as poly-L-lysine [8, 29, 34–36, 63], poly-D-lysine [20, 67], polyethyleneimine [43, 53], polyornithine [68], fibronectin [9, 10, 34, 61, 68, 70], laminin [17, 19, 35, 36], or collagen [60].
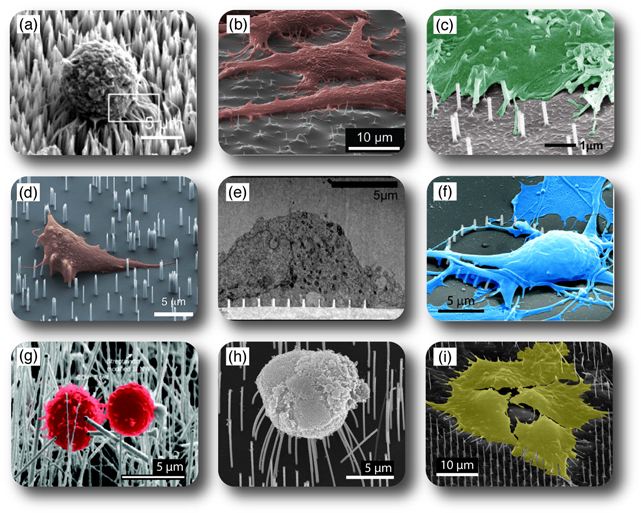
The cell types used in the different studies are asty 25 varied as the materials and geometries (figure 2). A wide range of typical model cell lines (e.g., CHO, HeLa, HEK293, and PC-12) have been successfully interfaced with many types of materials (carbon, silicon, gallium phosphide, indium arsenide, silica, aluminum oxide, zinc oxide and copper oxide), indicating a general biocompatibility and versatility of the NS arrays (table 1). Also, more vulnerable cell lines, such as primary neurons, primary immune cells, stem cells, and even tissue slices, have been successfully interfaced with NSs made from a variety of materials (e.g., carbon [34–36], silicon [5, 7, 8, 14, 16, 18, 20, 41, 44, 50, 51], platinum [29], gallium phosphide [17, 19, 21, 27, 54], quartz [63], zinc oxide [62], and iridium oxide [70]). One could imagine that by rationally combining certain platforms and cell types, the applicability of each system could be optimized.
Figure 2. Cells on arrays of NSs. (a) A549 cell on silicon NSs [Reproduced with permission from [28]. Copyright IOP Publishing.], (b) HeLa cells on copper oxide NSs [Reproduced with permission from [59]. Copyright 2013 Wiley.], (c) CHO or HeLa cell on hollow aluminum oxide NSs [Adapted with permission from [6]. Copyright 2012 American Chemical Society.], (d) natural killer cell on silicon NSs [Adapted with permission from [7]. Copyright 2012 American Chemical Society.], (e) cortical neuron cell on quartz NSs [Adapted with permission from [63]. Copyright 2012 American Chemical Society.], (f) cortical neuron cell on a ring of platinum NSs [Adapted with permission from [29]. Copyright 2010 American Chemical Society.], (g) T lymphocyte cells on silicon NSs [Adapted with permission from [41]. Copyright 2010 American Chemical Society.], (h) retinal cell on gallium phosphide NSs [Reproduced with permission from [27]. Copyright Elsevier 2013.], and (i) HEK293 cells on indium arsenide NSs (personal image).
Download figure:
Standard image High-resolution image3. Cell response to nanotopographies
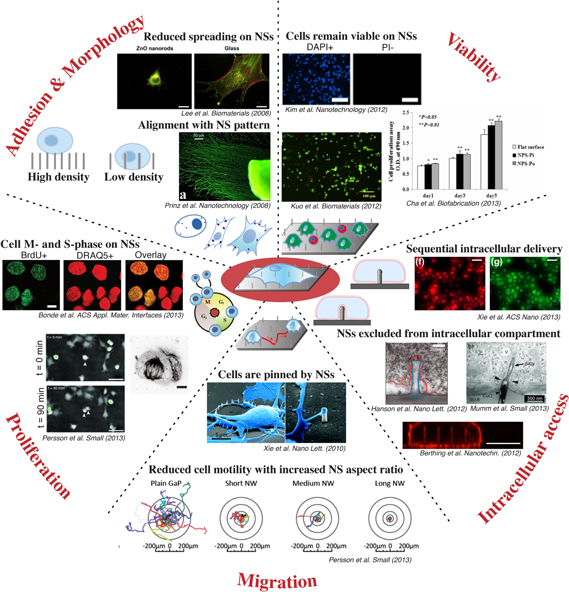
In vivo cells are exposed to a three-dimensional (3D) extracellular matrix with micro- and nanoscale components. Accordingly, cells in vitro are readily affected by both micro- [71–73] and nanoscale [15, 74, 75] surface topography. When an adherent cell comes in contact with a surface, many parameters will affect the continued fate of this cell (figure 3). It can respond to the molecular composition and surface chemistry of the material [76], its flexibility and deformability [77–79], and even small changes in the geometry and topography, such as the NS density [13, 14], diameter [43, 80, 81], and height [44, 55, 82, 83], are reported to affect the fate of cells. Furthermore, by modifying those properties, one can create both cell repelling- [61, 62, 65, 82–85] and cell promoting [13, 29, 46, 54, 65] surfaces. The cells' response is commonly monitored to confirm the biocompatibility of a material, but is also increasingly being exploited to design artificial materials that can induce desirable cell responses, such as promoting or reducing adhesion, assisting differentiation, or dictating cell spreading. Gathering information on how cells are affected by the interface with NS materials can establish a predictable correlation between surface topography and cell behavior. This information is highly valuable for the continued development of NS-based materials for cellular applications. For this purpose, systematic investigations serve a particularly important function, as allowing one topographical parameter (NS diameter, height, density, regularity, etc) to be changed independently may uncover the effect of each. Furthermore, using rationally designed arrays of ordered NSs ensures that each cell is exposed to exactly the same environment and thereby data variability is greatly reduced. In contrast, each cell on random arrays of NSs is in a unique situation, different from the other cells, and therefore, such a readout is heavily averaged across the surface and less precise.
Figure 3. Representative images of cellular behavior observed after interface with arrays of NSs. Adhesion and Morphology: On low-density NS arrays, cells are often seen to sink down along the height of NSs and generally adhere better than on high-density NS arrays, where cells often grow on top of the NSs (left illustration [Reproduced with permission from [13]. Copyright 2013 American Chemical Society.]). Cells show greatly reduced spreading on certain NS arrays (Lee et al Biomaterials 2008 [Reproduced from [61]. Copyright 2008, with permission from Elsevier.], scale bars: 20 μm), and commonly align with the NS pattern (Prinz et al Nanotechnology (2008) [Reproduced with permission from [21]. Copyright IOP Publishing.]). Bottom illustration: cells with different morphology, as well as different number and sizes of focal adhesion points (spots). Viability: Cells remain viable on many types of NS arrays, where most cells have an intact nuclear envelope (PI-) (Kim et al Nanotechnology 2012 [Reproduced with permission from [28]. Copyright IOP Publishing.], DAPI+ = all cell counterstain, scale bars: 200 μm), and maintained esterase activity (Kuo et al Biomaterials 2012 [Reproduced from [44]. Copyright 2012, with permission from Elsevier.], scale bar: 100 μm). In some cases, the viability is superior on NSs (NPS-Pi) compared to the flat control several days after seeding (Cha et al Biofabrication 2013 [Reproduced with permission from [64]. Copyright IOP Publishing.]). Illustration: living (green) and dead cells (red) on NS array. Proliferation: Cells are seen to divide and increase in numbers over time on a variety of NSs. More cells are seen to synthesize new DNA (BrdU+) on NSs than on controls (Bonde et al ACS Appl. Mat. Interfaces 2013 [Reproduced with permission from [13]. Copyright 2013 American Chemical Society.], DRAQ5 = all cell counterstain, scale bar: 10 μm), and able to make mitotic spindles (Bonde et al ACS Appl. Mat. Interfaces 2013 [Reproduced with permission from [13]. Copyright 2013 American Chemical Society.], scale bar: 5 μm), and divide into daughter-cells (Persson et al Small 2013 [Reproduced with permission from [55]. Copyright 2013 The Authors. Published by Wiley.], arrow heads in image center, scale bars 50 μm). Illustration: sketch of the cell cycle. Migration: Cells are pinned by NSs (Xie et al Nano Lett. 2010 [Adapted with permission from [29]. Copyright 2010 American Chemical Society.], scale bars 5 μm (left) and 2 μm (right)), and exhibit a reduced cell motility on arrays of NSs (NW) compared to flat control (Persson et al Small 2013 [Reproduced with permission from [55]. Copyright 2013 The Authors. Published by Wiley.]). Illustration: cell moving across an NS array. Intracellular access: Arrays of hollow NSs can repeatedly deliver material to cells, using for instance NS-based electroporation to deliver plasmids encoding fluorescent proteins (Xie et al ACS Nano 2013 [Adapted with permission from [67]. Copyright 2013 American Chemical Society.], scale bars 50 μm), but NSs are often excluded from direct intracellular access, as shown with both electron microscopy (Hanson et al Nano Lett. 2012 [Adapted with permission from [63]. Copyright 2012 American Chemical Society.], scale bar: 20 nm, NS = blue, cell membrane = red; Mumm et al Small 2013 [Reproduced with permission from [59]. Copyright 2013 Wiley.], scale bar: 500 nm) and fluorescence microscopy (Berthing et al Nanotechnology 2012 [Reproduced with permission from [57]. Copyright IOP Publishing.], scale bar: 10 μm). Illustration: a NS gaining direct intracellular access (top) or remaining enclosed in membrane (bottom).
Download figure:
Standard image High-resolution image3.1. Cell adhesion
Cell adhesion (figure 3) is mediated by large protein scaffolds known as focal adhesion (FA) points. These FA points are tightly associated with the actin cytoskeleton and together they control a range of cellular responses, such as morphology, migration, and adhesion, that cells use both for sensing and responding to their environment [86].
3.1.1. Adhesion as a function of NS density
Although some high-density (⩾30 NS/100 μm2) NS arrays are able to support the growth of adherent cells [51, 55, 63, 64], they are generally seen to inhibit cellular adhesion [42, 61, 62, 82–85], whereas medium- (>1<30 NS/100 μm2) and low- (≦̸1 NS/100 μm2) density NS arrays almost consistently support, and even promote adhesion [13, 29, 46, 74]. Kim et al demonstrated that a NS array was able to capture roughly 90% of the added cells, whereas the flat controls captured less than 25% [28]. The readiness of cells to detach from NS arrays has been investigated through assays involving either liquid flow or increasing centrifugal speeds, which, in agreement, demonstrate that NS arrays markedly reduce cell detachment [13, 46]. The borderline between NS densities being either adhesion promoting or reducing is somewhat indistinct, due to the different geometries and materials used to fabricate the NS arrays. However, a key difference between the high-density and low-density NS arrays is the surface area available to the cell. When approaching a very dense array of NSs, cells are forced to adhere directly to the NSs themselves, and are not able to reach the underlying flat surface. The cell is thus experiencing a reduced available contact area, which has been reported to reduce adhesion [87]. An early study reported that the cells preferably formed FA points at the NS locations [21]. However, in a focused study, we recently demonstrated that the cells do not adhere directly on the NSs themselves [13], but rather the FA points are formed on the surface between the NSs. The surface area of the NSs is postulated to simply be too small for adequate FA formation, as the cells are unable to adhere to NSs below a threshold of 70 nm either in height, diameter, or spacing [88]. Interestingly though, despite the fact that the NSs are not directly involved in the FA formation, the cells nevertheless show an up-regulated adhesion profile in their presence and form more and larger FA points when interfaced with a range of NS array densities [13].
The observed changes in cell adhesion on vertical NSs are suggested to be both an effect of modified FA point formation and a direct effect of the altered structural support. The topographic environment that is introduced to the cells by the NSs could allow a tighter, 3D-like interaction with the actin cytoskeleton [13, 29, 88]. This would in turn stabilize the membrane for both actin polymerization and FA point formation [16, 29], and consequently, offer a physical support that could influence cell adhesion on NS arrays.
3.1.2. Cell position along height of NSs
Cells are generally seen to grow on top of NSs arrays of higher density [27, 28, 37, 40, 41, 44, 46, 54, 60] and sink all the way down to the underlying surface at lower densities [20, 43, 57, 59] (figure 3). Though the phenomenon is apparent in high-density vs low-density parts of random arrays of NSs [48, 55, 57], it is most clearly illustrated in ordered arrays. Here, the cells' position along the height of the NSs is highly dependent on the NSs spacing, as the cells remain close to the top of the NSs in arrays with 0.5 μm spacing [63], and sink increasingly closer to the underlying surface, as the NSs spacing is incrementally increased toward 10 μm spacing [13]. A critical spacing of 2–3 μm between NSs is determined both experimentally and theoretically, above which the cells sink down to the surface, and below which the cells adhere on top of the wires [13, 14].
Naturally, the cells' position along the height of the NSs is dependent on the time elapsed since interfacing, where cells remain on top of NSs 15 min after seeding, but have sunk in between the NSs at 1 h [5]. It is also dependent on cell type (unpublished results), as HEK293 cells reach down to the support, whereas NIH/3T3 cells remain suspended on top of the NSs with the same spacing [4]. In an interesting study, the distance between the cell and the different parts of the NSs and underlying support has been determined based on focused ion beam (FIB) scanning electron microscopy (SEM) imaging [63], showing that the soma of cortical neurons is less than 50 nm from the underlying support.
3.2. Cell morphology
Cell morphology refers to the basic structure and appearance of a cell. When interfacing cells with NSs, the surface topography plays an important role in dictating the cell morphology (figures 2, 3). Micrometer-sized pillars [72, 89] and minute nanoroughness [15, 74, 75] can drastically alter the morphology of a cell. Cell morphology is a yardstick for normal cell function, but it also has a direct influence on gene expression [90] and cell survival [91], and can thereby affect any aspect of cell health, signaling, and differentiation. Cell morphology is commonly described as a qualitative estimation of cell appearance, but is increasingly being acknowledged for its importance through objective and quantitative investigations.
3.2.1. Qualitative estimation of cell morphology
Cell morphology on arrays of NSs is, in many cases, described in vague qualitative terms. In some cases, it is noted, without comparison to control surfaces, that the cell morphology remains normal or unaltered on NSs [4, 5, 7, 23, 36, 41, 60]. When taking vital controls into account, maintained cell morphology has been observed when comparing a NS surface with its corresponding flat control [9, 20, 34, 59, 63, 67], for instance, where it is seen that cells can spread normally despite the presence of a random array of hollow NSs [6]. Other studies have found the morphology of cells to be qualitatively different on NSs compared to controls [16, 27, 29, 38, 46, 51, 54, 67], for instance, mouse fibroblast cells have a greater variability in size and shape on gallium phosphide NSs [55]. However, this latter finding can be a consequence of comparing a confluent cell layer to single cells, where in the latter case, the cells are more free to reach out and able to spread.
3.2.2. Quantitative measurement of cell morphology
Quantitatively, parameters such as cell size and shape can be characterized in detail by using algorithms for cell area, aspect ratio, circularity, solidity, etc. This approach allows for objective and precise investigations of both cell soma-, axon-, and neurite properties [13, 14, 18, 28, 40, 44, 61, 64]. With few exceptions [64], cells are generally rounder with a smaller projected cell area on NSs than on the corresponding flat controls [13, 14, 40, 44, 61], and increasing the NS height leads to a further decrease in the cell area [44]. By using a NS array where the distance between NSs has been incrementally reduced, we recently demonstrated that the cell area becomes significantly smaller as the NSs become more tightly packed [13]. The morphological differences between cells on different NS spacings are noted as early as a few minutes to few hours after interfacing, i.e., when the cells are removed from their continuous culture and seeded onto the NSs.
3.2.3. Morphological maturation of cells
Differentiation of cells, especially neurons and stem cells, is of great scientific interest. Apart from the cell size, modifying NS spacing also affects cell elongation [13, 14], a maturational hallmark of many types of neurons. On NSs separated 2 to 10 μm apart, cells are more elongated with lower NS density, as compared to controls and higher densities [13]. Another study demonstrates that the critical distance between NSs that polarizes the cell extensions is around 2 μm, irrespective of the NS material, dimensions, and cell types tested. Elongating the NSs causes the cells to align even more with the NS pattern [14], a phenomenon also seen for arrays of random NSs [27]. This shows that cells can not only mature in spite of NSs [5, 8, 34, 53, 54, 60, 63], but that patterned high-aspect-ratio NSs can enforce maturation on their own and subsequent tuning of the NS array topography modifies the extent thereof. Cell maturation on NSs is also confirmed by non-morphological findings, showing that cells on NSs can start beating [9, 43, 70] and express markers for mature cells [14, 38, 44, 50, 51, 64].
Taken together, it is clear that even minute changes in NS array topography can have dramatic consequences for projected cell area, elongation of cells and their processes, and cell maturation. An interesting phenomenon, found in almost all reports, is that irrespective of cell type and NS material, geometry, density, and regularity, cell processes generally reach out toward the NSs; often, the cells arrange themselves according to the position of the NSs [13, 14, 16, 29, 67].
3.3. Cell viability and death
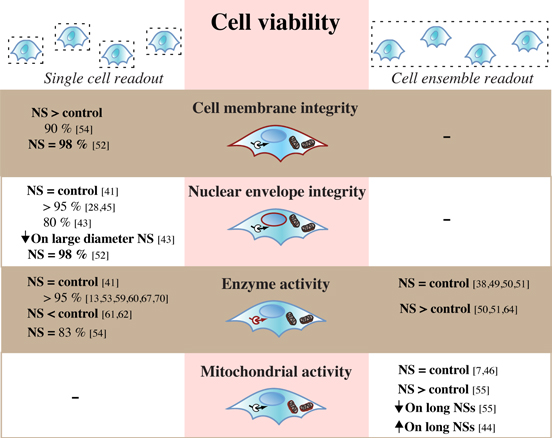
Once a cell has adhered to and spread on the NS surface, it is crucial to investigate the survival of the cells. If the cells become unhealthy, or even die, while interfaced with the surface, the platform would naturally be inadequate for biosensing purposes. The term cell viability (figure 3), or cell health, is used rather unspecifically, as there are many alternative ways to assess it (figure 4). Cell viability is, in many cases, observed indirectly by the mere presence of cells at a certain number of days following interface, assuming that dead cells would have detached by then. Cells have been seen to remain on NS arrays for at least 2–3 weeks [20, 22]. Several properties—such as the status of the cell membrane and nuclear envelope, presence of cellular enzymes, activity of the mitochondria, or synthesis of proteins—can define a healthy cell and therefore, there are many ways to measure the cell viability more directly. Moreover, the assays available can retrieve either ensemble- or single-cell based information, providing an average result for all cells on the sample or details on each individual cell, respectively.
Figure 4. Viability assays used to investigate cells on NS arrays—the output resolution, parameters, and results. Cell viability on NS arrays is assessed in a variety of ways and is predominately based on the integrity of cell membrane or nuclear envelope, as well as activity of intracellular enzymes and the mitochondria. Depending on the assay, the readout has a single-cell or cell-ensemble resolution. Percentages refer to the percent of viable cells given by the specified reference. 'NS = control,'NS < control' or'NS > control' = respectively same, lower, or higher viability on NS arrays compared to flat control surfaces. 'NS = X%' = viability provided only for cells on NS, not on control. Arrow pointing down or up = respectively reduced or increased viability. Numbers in square brackets correspond to numbers in reference list.
Download figure:
Standard image High-resolution image3.3.1. Integrity of cell membrane and nuclear envelope
A structurally intact cell is imperative for the maintenance of homeostasis and cell survival. One way to evaluate cell viability with single-cell resolution is to assess the physical status of the cell membrane and nuclear envelope (figure 4). In cells on NSs, a high viability (90–98% [52, 54]) has been measured based on the status of the cell membrane alone. Instead investigating the nuclear envelope, Kim et al 2007 found a good cell viability (80%); however, an increased NS diameter had a clearly negative influence [43]. More recent investigations have estimated cell viability to be even higher [41] (>95% [28, 45, 52]) on both Si and quartz NSs. Although a high viability is favorable, the detected viability levels are most importantly similar, or even superior [59], on the NSs compared to the corresponding flat controls. This shows that, in general, thin NSs (diameter <200 nm) do not impair the cell membrane or nuclear envelope, possibly because of the membrane's ability to bend and conform to the NSs [9, 57, 59, 63] (figure 3).
3.3.2. Activity of cell enzymes
Some assays for cell viability take advantage of the presence of intracellular enzymes that are only active in healthy cells (figure 4). Using this type of assay as the single readout, a high level of cell viability (80%) has been observed on NSs [54]. However, measurements of cell enzymes are commonly combined with assays to investigate the nuclear membrane as well [7, 10, 20, 44]. With this approach, we and several other groups have shown that the cell viability is commonly similar between NSs and the control [6] (>90% [13, 53, 59, 60, 67, 70]), although a few reports show a reduced viability on NSs compared to a flat control [61, 62]. The activity of cell enzymes can also be evaluated through an ensemble-based readout, which has shown either similar viability on both NSs and controls [38, 49–51], or a significantly elevated viability for cells on NSs compared to flat controls [51, 64].
3.3.3. Activity of mitochondria
The metabolic activity of mitochondria is the foundation for several ensemble-based viability assays (figure 4). With these techniques, it has been shown that the viability of cells is unaffected by the presence of NSs [7, 46]. Increasing the NS height influences the cell viability; however, changing this NS property can, according to conflicting evidence, result in either increased [55] or decreased [44] mitochondrial activity.
3.3.4. Apoptotic cell death
The methods previously described for investigating cell viability do not distinguish between the specific types of cell death that may be occurring. By using single-cell based approaches, cell viability investigations have been combined with markers specific for apoptotic cell death. This has revealed very low [27] (<1% [62]) levels of apoptosis on NS arrays, although transiently higher levels (10–20% [5]) have been detected. In a study that included a control, it was shown that few cells underwent apoptosis on the NSs (<5%), whereas a very high number of apoptotic cells were detected on the flat control surface (>70% [51]), highlighting that the presence of NSs can enhance cell viability.
3.3.5. Cell stress and protein expression
Other methods have been used to comment on cell viability on NSs. Cell stress levels have been measured and increasing levels of reactive oxygen species [55] were detected as the NS length increased. Analyzing the expression of mRNA and proteins demonstrated similar levels of house-keeping genes [5] and genes associated with stress [6], as well as expression of both cytosolic and membrane proteins [53] in cells on NSs and flat controls. Also, the activity of inflammatory markers [7] has been investigated, showing no activation of the innate immune system on NSs.
Finally, it is apparent that many approaches are available to investigate cell viability and death (figure 4). Each readout must be valorized according to the type of information it provides, i.e., ensemble-cell or single-cell resolution. Ensemble-based assays are generally analyzed in a plate format, and provide a faster readout that represents an average of all cells across the entire surface. The single-cell based assays are evaluated using microscopy, and they can thereby give more detailed information on cell viability and simultaneously provide other types of readout that can be correlated, such as cell size and morphology, and the position of the NSs. As the assays evaluate cell viability from several perspectives and in varying detail, choosing different approaches can cause dissimilar results. Encouragingly though, for the development of NS arrays for cellular purposes, the reported viabilities are usually as high on the NSs as they are on the corresponding flat controls.
3.4. Cell proliferation
The ability of cells to proliferate on NSs is important to consider both as a further measure of cell health, but also with respect to the types of applications the NS array can be used for (figure 3). The term proliferation is often used in a careless manner, through general statements that cells grow, divide, and proliferate on NSs, without providing further data [5, 10, 22, 23]. Both adhesion and viability are to some extent part of the term proliferation, which should more precisely be defined as both cell division and cell number increase. The combination of the two is necessary, as observing a lower relative number of cells over time on NSs, compared to flat controls, does not necessarily correspond to a lower division rate on NSs, as each method on its own do not account for factors such as cell detachment and death.
3.4.1. Cell division
The cell cycle progression on NSs has been investigated using methods on both fixed and living cells. Using immunocytochemical labeling of cell cycle markers, it has been shown that a cell cycle antigen is present, to a similar extent, on both the NSs and the control [27], and that more cells are in the cell cycle S-phase on NSs than on the corresponding control [13]. As using these types of markers does not exclude the possibility that the NSs modify the cell cycle by increasing or decreasing the dwell time in each phase, it is useful to combine these methods with investigations of cell division in live cell samples to follow the same cells for extended periods. Using this approach, we have along with others, demonstrated that cells can undergo cell division on arrays of both vertical and semivertical NSs [4, 6, 13, 16]. In contrast, cell division has been reported to be disturbed on GaP NSs, where cell division is either aborted or aberrant multinucleated cells are created [55].
3.4.2. Cell number increase
To investigate whether an increased or decreased rate of cell division results in a net change of cell numbers, i.e., whether the cell population is actually proliferating, cell division must be combined with investigations of the total numbers of cells over time. In some cases, a change in cell number on NSs is stated without actual data [5, 37, 67], whereas in others studies, the change in cell number is followed for up to seven days [4, 7, 37, 38, 46, 55, 64]. Based on ensemble assays, the cell numbers are, over time, either maintained [46] or increased [64] on NSs compared to controls. By microscopically counting cells at different time points after interface using time-lapse imaging, cell numbers are seen to increase over time on both NSs and controls, but to a lesser extent on the GaP [55] and Si NSs [39]. In comparison, cell numbers decrease on both ZnO NSs and the control, but to a greater extent on the NSs [62]. Cell numbers have also been shown to increase [4] or decrease [7] at similar levels on NSs and controls.
In conclusion, only a few studies have thus far have compared the level of cell division between NSs and control surfaces, and the investigations for cell number increase show wide discrepancies. Therefore, no general tendencies can currently be distinguished when it comes to cell proliferation on NS arrays. Following both cell division and the change in cell numbers over time also considers the level of cell death and detachment from both NS and control surfaces, and thereby, this strategy provides a highly valuable understanding of cell proliferation on NS arrays.
3.5. Migration of cells
The migratory pattern (figure 3) has been investigated for primary rat neurons interfaced with an array of ordered platinum NSs. By positioning NSs in a circle, it was demonstrated that the cells remained virtually stationary over a four-day period, while cells on the flat control surface were mobile and migrated up to several hundreds of micrometers during the same period of time [29]. This finding has also been confirmed on both random and ordered arrays of NSs, where cells remain stationary on solid [13, 55] and hollow [6] NSs, whereas they migrate longer distances on flat control surfaces [6, 55]. The reduced migration might be dependent upon surface flexibility, as the cells migrate less on longer NSs rather than on shorter ones [40, 55].
When summarizing all reported cellular responses, it is clear that interfacing with arrays of NSs substantially affects the cellular fate (figure 3). To be able to correlate surface topography with cell responses represents a leap forward for the rational design of ordered arrays of NSs for biological applications. The endeavor is challenging, as the experimental conditions are close to unique for each study in terms of NSs (NS material, shape, diameter, height, array density, regularity, etc), as well as the variability in cell types (table 1) and control surfaces chosen. Interpretations of the results are further complicated, as different conclusions to the same question are drawn based on different types of assays and measurement strategies (figure 4). Furthermore, even with the same experimental setup, assays with different resolutions are seen to provide contradictory results. In a recent study, the single-cell count assay and the ensemble-based Alamar Blue assay—used by others to evaluate the number of cells on a surface [44, 46]—showed opposite results, despite the same combination of NS arrays and cells used [55]. For many of the cell responses it can be very useful to make detailed quantitative measurements, as even small changes of the experimental conditions can have large consequences on the cell fate. In addition, making repeated measurements over time can ensure that the observed response is lasting and is not influenced by the seeding process or the potential cell synchronization following cell interface. The cell responses are more clearly identifiable in systems with well-defined topographies, instead of random arrays where the cell response can vary across the surface. Interestingly though, it seems that several properties can be generalized to a number of systems, i.e., combinations of NS characteristics and cell types. Almost all systems tested support cell adhesion and viability; although, there seems to be a greater risk for cell death and detachment in systems with very high NS density. Cells are generally rounder with a smaller projected cell area on NSs than on flat controls. Although not creating FA points directly on the NSs, it seems cells prefer to adhere to NS arrays, rather than on flat controls, and arrange their adhesion in relation to the NS pattern. Accordingly, cells are captured on NSs and migrate less than on the flat controls.
The many interesting NS-induced cellular responses might be the result of a modified gene and protein expression. This can indirectly be the outcome of remodeled FA signaling complexes [91], possibly in conjunction with adapted levels of intracellular calcium [51], or as a result of the geometric constrains that the NSs introduce to the cells [90, 92]. It can also be a direct effect of the increased NS-induced nuclear membrane curvature, which increases the proximity between chromosomes and the nuclear membrane, and alter the gene expression [93].
4. Obtaining and maintaining intracellular access of NSs
To obtain access to the intracellular compartment is a major focus for the continued development of NSs for cellular applications. Naturally, for some applications, an intracellular access is of no, or little, importance. However, for many promising applications, intracellular access is crucial. This section will review methods that have been used thus far to obtain intracellular access of NSs [figure 5(a)], as well as the methods used to evaluate this access [figure 5(b)]. In many studies, the intracellular access of NSs is measured by expression of a protein coded by a delivered DNA plasmid. For ease, this readout will be referred to throughout as 'DNA expression.'
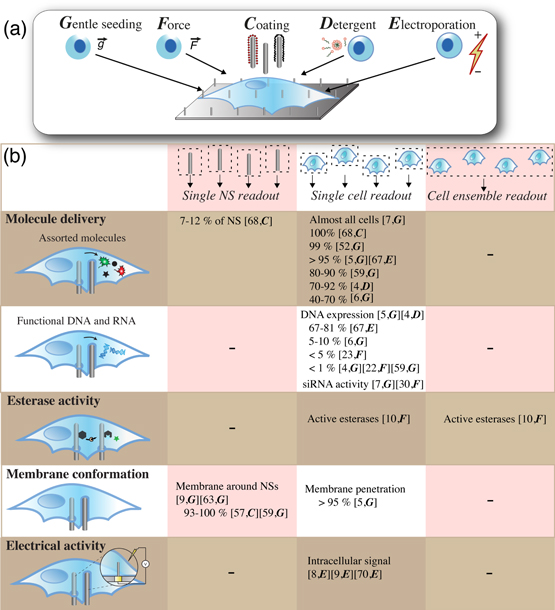
Figure 5. Alternative ways to interface cells with NSs and methods to evaluate their intracellular access. (a) Cells are often interfaced with NS arrays through gentle seeding (G), but a variety of directed approaches have been used to improve intracellular access. External force (F) can be applied through pressing or centrifugation, NS surface coating (C) may be adjusted, detergents (D) can be added to the cell medium, and cells have been exposed to electroporation (E). (b) The intracellular access of NSs is commonly judged by the delivery of small fluorescent or nonfluorescent molecules; by delivery of functional siRNA or DNA; by monitoring esterase activity on NSs; by investigating the membrane conformation around the NSs; or by electrical recording of distinguishable extra- and intracellular signaling profiles. Provided percentages refer to the percentage of cells containing or expressing a delivered molecule or the percent of NSs with or without intracellular access, as specified. Depending on the assay used, the results will have either single-NS, single-cell, or cell-ensemble resolution. In square brackets: numbers correspond to numbers in the reference list and capital letters in italics (G, F, C, D, E) correspond to the interfacing approaches defined in a. '-' = an option that has not been evaluated.
Download figure:
Standard image High-resolution image4.1. Intracellular access of NSs following standard culture
NSs were initially believed to gain intracellular access spontaneously when interfaced with cells through standard culture methods [figure 5(a)]. Access was evaluated based on electron microscopy and confocal fluorescence microscopy of NSs excluding a volume of cytosolic dyes [37, 43, 54], as well as DNA expression, although with very low efficiency [43] [figure 5(b)]. This is also the conclusion from a later report, where both the NSs and plasma membrane were fluorescently labelled and imaged with confocal microscopy, showing that most cells were impaled by the NSs within 1 h after seeding [5, 7].
However, experimental evidence eventually suggested that spontaneous access is not always gained, possibly only for as few as 1–10% of the NSs [6, 68]. With a detailed single-NS resolution, we used fluorescence labeling of a membrane protein [figures 3, 5(b)] to show that the plasma membrane generally enclosing the NSs, irrespective of the wide range of NS heights and densities investigated [57]. The method developed was used by Mumm et al on a platform with other types of NSs and cells, confirming that the NSs remain wrapped in cell membrane also under these conditions [59]. Nevertheless, highly efficient intracellular delivery of small fluorescent or nonfluorescent molecules is still reported with both solid [5, 7, 52, 59] and hollow [6] NSs [figure 5(b)]. A high delivery efficiency, however, is likewise observed on flat control surfaces [52, 59], indicating that delivery of fluorescent molecules might not be directly associated with NSs penetrating the cell membrane, but rather assisted by the local up-concentration of molecules at the NS-cell interface, or by defects at the highly curved membrane that are too small or transient to be detected with a membrane labeling technique. Either way, this stresses the importance of including proper controls when using the delivery of molecules to evaluate intracellular access. Delivery of functional siRNA or DNA is also reported, but with generally lower efficiencies than for the previously mentioned small molecules. This shows how the delivery of certain molecules does not necessarily equate delivery of especially larger molecules or molecules that require maintenance of functionality. Furthermore, several studies using transmission [59, 63] and scanning electron microscopy following focused ion-beam milling [9, 48, 55] have investigated the interface between NSs and the cell membrane. These techniques also reveal that the plasma membrane wraps tightly around the NSs [9, 59, 63] (figure 3). Using a recently developed theoretical model, we actually predict this behavior, because in many cases during standard culture, the cells can energetically afford to simply bend the membrane around the NSs [13], giving them no incentive to be penetrated. However, a very recent theoretical study proposes a post-deformation, adhesion-mediated mechanism for spontaneous penetration and explores the probability of such intracellular access as a function of NS array dimensions and mechanical properties of the cells [94].
4.2. Assisted intracellular access of NSs
As intracellular access is not spontaneously acquired for many platforms, alternative strategies are developed to obtain intracellular access and to further increase the number of NSs gaining access [figure 5(a)]. One strategy evaluated is to modify the surface coating of the NSs. By increasing the nitrogen content of vertically aligned carbon nanofibers, they are able to retain more DNA, and subsequent interface with cells was shown to yield a higher number of DNA-expressing cells [22]. Interestingly, modifying the NSs with aminosilane increases the number of NSs gaining a direct intracellular access from 0–7% [57] compared to bare and polyethyleneimine-coated NSs [figure 5(b)]. Furthermore, by coating NSs with other adhesion promoters, the Melosh group recently demonstrated that polyorthinone and fibronectin both increase the number of NSs with cytosolic access, as well as significantly affecting the penetration kinetics, thus making the direct cytosolic access a quicker and more likely event [68]. The same group has also developed an approach to maintain the direct access of NSs to the intracellular compartment, where the addition of a hydrophobic ring around the NS increases the adhesion strength to an artificial bilayer [95, 96] or to the membrane of a red blood cell [97]. The relevance of defining a hydrophobic region to assist membrane penetration has also been confirmed by modifying other types of NSs with a phospholipid bilayer [98].
Applying force to the cells during interfacing is one of the earliest tactics applied to enhance intracellular access [22, 23, 30, 32, 33] [figure 5(a)]. It has been shown that centrifuging cells onto carbon NSs is not enough for efficient delivery of DNA. Inverting the NS array on top of the cells and subsequently pressing or tapping it with tweezers somewhat improves the delivery efficiency but also introduce large variations in delivery efficiency across each array [22, 23, 32, 33] [figure 5(b)]. In an interesting study, outside the scope of this review, the force needed to penetrate the plasma membrane was probed using a single NS on an atomic force microscope tip; subsequently, the short-term viability of the penetrated cells was also investigated [99–101].
A recent strategy that to some extent also includes forcible penetration has been denominated the 'sandwich' assay. Here, the cells adhere to one NS array that is inverted and placed on top of another NS array (the probe array) functionalized with enzyme substrates. Intracellular access is either shown by a fluorophore on the substrate, which is then cleaved with intracellular enzymes and left behind in the cytosol, or by examining the probe array using mass spectrometry to verify substrate cleavage, thus showing retroactively that the substrate has been in contact with intracellular enzymes [10] [figure 5(b)].
Detergents are commonly used in immunocytochemistry to deliver the large antibodies across the membrane of fixed cells. By using a mild detergent solution to destabilize the membrane [figure 5(a)], Peer et al show that macromolecules are able to enter the cell through hollow NSs, while still maintaining cell viability [4]. The direct intracellular access of the NSs is shown through delivery of a fluorescently labelled molecule and a DNA plasmid encoding a fluorescent protein from a reservoir below the hollow NSs. To achieve delivery, it is necessary to both have NSs present (pores without NSs do not deliver cargo) and a low concentration of a detergent in the solution. This demonstrates that a combination of mechanical strain and detergent can facilitate the intracellular access of NSs.
The cell membrane can likewise be destabilized using electroporation [figure 5(a)]. Nanoelectrodes are small, but tightly coupled to the membrane, and they can thus create a strong electric field using only a small voltage to increase the cell membrane permeability transiently and locally. Using such NS-based electroporation, direct intracellular access of NSs has been demonstrated by several groups. The shape of the measured action potentials shows when access is gained and it is demonstrated that NSs can gain access to the same cell repetitively over several days [9, 70]. In a different system, access is spontaneously gained to more than half of the cells, and access can be obtained for the rest through electroporation [8]. In a platform combining electroporation and relatively thick (250 nm diameter) hollow NSs to avoid any spontaneous penetration, Xie et al show that electroporation is necessary to obtain intracellular access of the NSs. The access is similar to previous reports shown by delivery of a fluorescent dye and DNA plasmids encoding fluorescent proteins through the hollow NSs with high efficiency [67] [figures 3, 5(b)]. Intriguingly, all of these electrophysiology-based systems [8, 9, 67] congruently reveal that the intracellular access of the NSs is transient, on the order of minutes, before the membrane reseals.
In conclusion, a variety of methods has been used to interface cells with NSs [figure 5(a)] that, with variable success rates, have assisted the intracellular access of NSs. Modifying the NS surface coating increases the number of NSs gaining direct intracellular access by up to seven percentage points [57, 68]; by applying force, the number of cells expressing delivered DNA increases to at least 5% [23]; both detergent solutions and electroporation provided high efficiencies of molecule delivery (70–95% [4, 67]) and DNA expression (70–80% [67]), as well as a major improvement in the number of NSs with direct electrophysiological access to the intracellular compartment [8, 9] [figure 5(b)]. As previously shown, different conclusions are drawn regarding the ability of NSs to spontaneously gain intracellular access [figure 5(b)], which can have several explanations. Naturally, the NS access may differ when different cell lines are presented with the same topography, or when one single-cell line is presented with different topographies. Therefore, the dimensions and positioning of the NSs should ideally be controlled to promote consistency between experiments. Furthermore, even though there is a significant difference between the delivery efficiency measured per NS or per cell, i.e., the intracellular access of NSs versus molecule delivery efficiency, the two concepts are commonly made equivalent. Thus, when discussing intracellular access of NSs, one should remember that the situation might not be identical for all NSs in a given sample. It is likely that any platform and interfacing method will result in some NSs gaining intracellular access, whereas others are wrapped by the plasma membrane. The method used to evaluate intracellular access may or may not allow estimation of the share of NSs that gain access. Some methods, such as membrane labeling and electrophysiology, can offer single-NS resolution, whereas others, such as molecule delivery, offer only single-cell or even ensemble-based resolution [figure 5(b)]. Finally, as illustrated by the electrophysiology-based systems, NS access can be transient and vary over time, thus also making the time point of observation important to consider.
5. Applications
The large difference in diameters between NSs and cells unlocks a wide range of potential applications for NS arrays in cellular investigations. Where other methods may suffer from low transfection efficiencies or poor survival subsequently, thin NSs are hypothesized to deliver material to many types of cells, many cells simultaneously, and potentially to different subcellular compartments without harming the cells. This could allow sensing and modifying activity within cells, or within a cellular network, and thereby provide unprecedented spatiotemporal information on biological processes. The NSs themselves could also provide this type of information owing to their innate electrical properties or be used directly to create patterns for cell network guidance and force measurements.
Converting the potential of NSs into real-life applications is still a task heavily pursued, and establishing NS arrays as a superior choice over competing methods is yet to come. Despite the struggle with technical details, NS platforms are becoming increasingly well characterized and several important proofs-of-principle have been brought forward in recent years.
5.1. NSs for molecular delivery
Cell membranes represent formidable barriers for delivery of exogenous macromolecules, which allows cells to maintain their integrity. To overcome this barrier is a major challenge in biological and medical research, where it is of great interest to deliver a broad diversity of molecules to study basic cell physiology and pathology and develop drugs to counteract disease. Traditional molecule delivery methods such as electroporation, viral vectors, and standard transfection reagents can be detrimental or toxic to cells, yield inconsistent transfection efficiencies [102], and generally require large volumes of precious biomolecules. In many standard cell lines, decent transfection efficiency can be achieved using standard transfection reagents. Although cells can be efficiently transfected with standard methods while in contact with NSs [53], using the NSs per se for this purpose can be a useful alternative. When using NSs for delivery [figure 6(a)], it has been demonstrated that these transfection efficiencies can be matched or even surpassed [figure 5(b)]. Several fluorescent and nonfluorescent molecules, such as siRNA, peptides, proteins, and DNA sequences have been delivered into cells with efficiencies ranging from 40–99% [5–7, 52, 59, 68]. The delivered molecules sometimes appear to remain enclosed in cytosolic vesicles though, which could explain why the subsequent efficiency of DNA expression often is well below 1% [22, 43, 59], and rarely up to 5–10% [6, 23] [figure 5(b)]. High expression efficiencies (70–80%) are thus far only achieved using a combination of hollow NSs and electroporation [67]. The role of the NSs in small-molecule delivery has been questioned, as recent evidence shows that high delivery efficiencies are, in some cases, also attained by the flat controls, in the absence of NSs [52, 59]. Encouragingly though, Chan et al demonstrated that although both NSs and flat control surfaces provide high delivery efficiencies, only by using the NS arrays would the delivered cargo avoid ending up in intracellular endo- and lysosomes [52].
Figure 6. Applications of arrays of a vertical 1D NS. NS arrays are generally designed along four lines of potential applications: molecule delivery (a), cell diagnostics (b), electrophysiology (c), and cell guidance (d). (a) A wide variety of molecules is delivered into cells using NSs, such as fluorescent dyes (green, yellow, red), DNA and siRNA (blue helices), and others such as peptides, proteins, and other bioactive molecules (black shapes). (b) Intracellular processes, such as enzyme activity (left insert) and the state of cytoskeleton (right insert) are probed using NSs. (c) Arrays of NSs are used to measure electrophysiology-based cellular activity. (d) By designing certain patterns, NS arrays can be used to guide cells and create artificial cell networks.
Download figure:
Standard image High-resolution imageOther cell types, such as certain primary cells, resting cells, suspension cells, and stem cells, have proven considerably harder to transfect [103, 104]. These cells are highly interesting in cell biology, but present an additional challenge, as they are resistant to even optimized transfection methods and either remain nontransfected or die during the process. Here, NSs have proven useful as promoters of exogenous molecule delivery into different types of primary immune cells.
A drawback of traditional transfection methods is that they generally do not allow for continuous or sequential delivery of molecules into the same cells. By using hollow NSs connected to an underlying reservoir instead [6], molecules can be delivered repeatedly into more than 70% of cells through passive diffusion in a detergent solution [4]. In a similar approach, hollow NSs were combined with localized electroporation, which demonstrated an on-demand, repeatable delivery to more than 95% of the cells [67]. The latter approach also demonstrated the highest NS-based transfection efficiency noted thus far, where the delivered DNA plasmids were expressed in up to 80% of the cells. These approaches are superior to microinjections, as they are done in an easy, high-throughput manner with benign manipulation of many cells simultaneously.
5.2. NSs as diagnostic probes
In addition to potentially modifying cell activity, molecules for detecting cell activity could also be delivered into cells [figure 6(b)]. The group of Nakamura is a major player in this field, where single NSs are used to measure enzymatic activity [105], deliver DNA molecules [106, 107], and investigate the status of the cell cytoskeleton [11, 12]. Recently it was demonstrated that this concept could be up-scaled to an array format, where the intracellular activity of three different enzymes was investigated. The enzymatic substrates were bound to an NS array, which was introduced into cells. After retraction of the NS array, the enzyme activity was evaluated based on intracellular fluorescence and mass spectrometry of the NSs [10]. This is an optimistic development toward real-time detection of cellular activity using NS arrays.
5.3. NSs as electrodes
The most common method for obtaining an electrophysiology-based readout of cell activity is using micrometer-sized patch-clamp probes. This technique is generally laborious, low-throughput, stressful to cells, and suffers from limited recording times [108, 109]. To upscale the method, micron-sized mushroom-like probes were demonstrated to enable intracellular-like neuronal measurements in an array format [110]. This work was soon followed by two exceptional demonstrations of intracellular label-free recordings of cell activity based on arrays of vertical NSs [8, 9] [figure 6(c)]. In these studies, pads of positioned vertical NSs were used to electroporate and measure the electrophysiological activity of cells. It was demonstrated that NS-mediated current injections could evoke action potentials in primary neurons [8], and that the action potential activity could be followed in the same cells over an unprecedented period of several days [9]. Further support for the use of NSs for electrophysiological purposes is that, as the membrane seals tightly around the NS, megaohm [8] to gigaohm [97] seal resistances are generated, which thereby greatly minimize the signal loss for NS-based electrodes. Recently, it was demonstrated that by using hollow instead of solid NSs, the cell-electrode coupling improved the signal even further and allowed for much longer recordings [70].
5.4. Applications of extracellular NSs
Arrays of vertical NSs are also developed for applications where the intracellular access of NSs is not imperative. The mere presence of NSs can physically influence the cells and thereby guide cells to form a network [figure 6(d)]. This has been demonstrated for neuronal cells, that either followed on top of [20, 21] the NS pattern or in between the NSs [19], thus creating artificial neuronal networks. The material, height, and diameter provide the NSs with a certain flexibility, and the forces exerted by cells have been measured by quantifying the deflection of individual NSs [16–18]. As described earlier, cells remain close to stationary on NSs [29, 55], providing the opportunity to use NS arrays to capture cells and to follow the same cell for extended periods of time. Additionally, because arrays of NSs influence the developmental fate of cells and allow growth of many specialized cell types, NS arrays are also considered for uses in tissue engineering and regenerative medicine [14, 38, 44, 50, 64, 111] and for their appropriateness as implants [27, 62]. Arrays of vertical NSs have moreover been proven useful for acellular applications, such as protein arrays [112, 113].
6. Conclusion and perspective
As is evident, there are many prospective biological applications for arrays of vertical NSs, and the numerous proofs-of-principle published recently illustrate the large toolbox that is currently being created in this advancing research area. The field is still in its infancy as, optimistically, many interesting novel NS arrays are even now appearing, and much effort is yet put on the basic characterization of their interface with cells. The lack of a unified nomenclature and a highly variable level of detail by which the NS interface and cellular responses are described are additional signs that the field is in an early stage of development, and possibly also reflects its interdisciplinary nature.
A few general trends and conclusions from combining cells with NSs are beginning to crystallize, whereas other results are clearly dissimilar. The discrepancy between some results reported thus far, notably in the case of NS access to the interior of the cell (figure 5) and cell responses to NS arrays (such as adhesion, viability and proliferation) (figures 3, 4), can be explained by a number of factors. In addition to the NS geometry (shape, diameter, height) and density, which have already been proven to influence the results obtained [13, 14, 43, 55], cells could perceive the various kinds of materials and coatings differently (e.g., toxicity) and the NS regularity in ordered versus random arrays. In addition, the large number of cell types used (table 1), each with its own subset of unique properties and thus expected distinct behavior (adhesion, spreading, resistance to transfection, etc), could explain discrepancies in results between different platforms.
In addition to the inherent variability of results obtained from combining various NS arrays and cell samples, discrepancies in results can also arise from different sample preparation and handling. For instance, cell confluency or cell fixatives and dehydration protocols can influence cell morphology [37] and studies in living cells and observations over time will also impact cell behavior unless a proper environment for cell culture is maintained. Moreover, results obtained on cells that are no longer in contact with the NS arrays might impact the results and conclusions on cell viability or other cell responses. Furthermore, in the context of cell viability and intracellular access of NSs, one should also consider the nature of the readout that is compared. It is essential to distinguish between different assays, and only compare results with the same level of resolution; e.g., the single-NS level, or the level of an entire cell population (figures 4, 5). Finally, given the intrinsic heterogeneity of a cell population (cells at different stages of their cell cycle and with various protein contents at a given time point), a sufficiently large cell population is required to allow comparisons of statistical relevance. These factors can potentially modify or cloud the interpretation of the cellular responses or intracellular access of the NSs; both scenarios can separately or in combination cause the discrepancies noted in the community. This suggests that a broader overview and a better understanding of some phenomena is attainable in the future, when the results described are better correlated to a careful description of the NS arrays and relevant control surfaces. Despite the fact that many different types of NS platforms have been characterized thus far, often unique in their combination of variables (material, topography, cells, methods), the obtained results are encouragingly similar in many cases despite the many different experimental conditions.
Some technical bottlenecks remain to be overcome by a part of the community. The difficulties in handling and imaging of arrays, together with the fragility of the NSs and cost of production, in some cases, may limit the development of NS arrays for various applications. These obstacles will most likely be overcome in the near future thanks to technological developments toward chips with well-defined and reproducible patterns. Such ordered arrays are especially important for ensemble-based readouts to ensure that all cells encounter the same environment. The manipulation of arrays for their access to the cell interior is still a challenge that will likely be solved in the future, owing to improved NS array design and coating, as well as optimization of sample handling and interfacing methods.
Even though the development in many cases is at an early stage, the rate and quality of the progression is very promising; the applications are surely to diverge further and novel applications might yet emerge. Before long, the potential of NS arrays could be converted into verified methods to measure and modify signaling pathways in living cells, cell diagnostics, and drug discovery. In conclusion, there is clear motivation to continue exploring NS arrays for cellular applications and establish the use of vertical 1D NSs as a superior choice over other current techniques.
Acknowledgements
The authors thank Peter Fuerst and Jesper Nygård for fruitful discussions. For financial support, we thank the Danish Agency for Science Technology and Innovation (The Danish Council for Strategic Research—CLIPS and ANaCell projects and The Danish Natural Science Research Council—FTP 11-116984), UNIK Synthetic Biology (funded by the Danish Ministry for Science, Technology, and Innovation).