Abstract
The sequencing of the human genome offered a glimpse of future medical practices, where information retrieved from the genome could be harnessed to inform treatment decisions. However, making DNA sequencing accessible enough for widespread use poses a number of challenges. This perspective article traces the progress made in the field so far and looks at how close we may be already to real-life applications.
Export citation and abstract BibTeX RIS
It has been only a little over a decade since the first sequencing of a human genome was completed (Lander et al 2001). Since then, the cost and time to sequence de novo an entire genome have come down considerably (Schloss 2008), and yet genome sequencing is still not cheap enough to allow personalized medicine. The sequencing of individual genomes rapidly (within a day or so) and economically (for less than $1000) could positively affect individuals' health via targeted drugs, and the assessment of likely future diseases.
The feat is clearly not easy. The four DNA bases—spanning a cross section of about 1 nm2—differ little from each other both chemically and physically. In addition, to complicate matters further they are linked to the same sugar–phosphate backbone. As a result, in order to distinguish them individually, we need a detection apparatus with nanometre-scale resolution (Zwolak and Di Ventra 2008).
Keeping costs at a minimum poses a number of requirements. (i) The fabrication of the apparatus needs to be relatively cheap. (ii) The DNA should not require any chemical preparation, amplification or labelling. (iii) We should be able to sequence considerably large chunks of DNA at a time, that is, at least gene size (104–105 nucleotides) if not the whole genome in one shot. For comparison, today's electrophoresis has a read length limit of about 1000 bases. Finally, (iv) the reading of the DNA bases from the sequence needs to be very fast.
Regarding points (i)–(iii) above, nanopores/nanochannels—namely holes of nanoscale dimensions in biological membranes or solid-state layers—have become the apparatus of choice. The sugar–phosphate backbone is charged in solution, so the DNA can be electrically driven through the pore. Some biological nanopores, such as α-haemolysin, come naturally equipped with the 'right' size to fit DNA strands (Branton et al 2008). On the other hand, solid-state pores can be made in practically any shape and size (Zwolak and Di Ventra 2008) thus offering added flexibility in the design of a sequencing machine.
The last point (iv) deserves a little more attention. It has become clear that electrical means hold considerable advantages. In particular the resolution of electrical techniques is as good as the cross section of the detection apparatus, which can be made as small as a few atoms wide (Zwolak and Di Ventra 2008). However, the question arises as to what type of electrical signal affords the fastest and most accurate detection scheme. So far, of the available options, the early suggestion (Kasianowicz et al 1996) of measuring ionic currents has been developed the most, and for obvious reasons. Once a nanopore is large enough to allow a single-stranded (ss-) DNA to translocate through it—but not too large so that only one base at a time blocks the flow of ions during translocation—it is enough to measure such currents and assign their value to each base.
Despite the simplicity of detection using ionic currents, the scheme suffers from some serious limitations. For example, the differentiation of each base has been demonstrated only by letting one base at a time pass through the pore hole. This requires either literally stopping (or considerably slowing down) the DNA translocation precisely when each base enters the pore (Manrao et al 2012), or cleaving the single nucleotides from the DNA strand sequentially using an exonuclease enzyme in close proximity to the pore entrance (Butler et al 2008). In either case, the read out of the current is much too slow (of the order of tens or hundreds of ms) and does not even compete with the methods already used for sequencing (Schloss 2008). Note there are two reasons why this time limitation is not easily overcome. First, the readout is achieved by detecting ionic currents, so the bases must dwell for a sufficiently long time at the pore entrance to collect enough ions to give a statistically meaningful signal. Second, the DNA speed is controlled by slow chemical reactions we borrow from Nature.
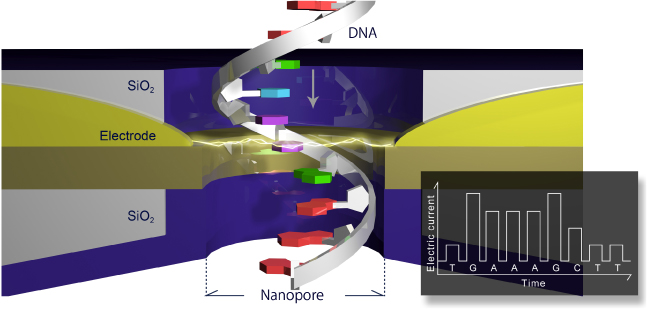
Instead, a different approach has been suggested (Zwolak and Di Ventra 2005) that requires the measurement of electrical tunnelling currents across the ss-DNA as it translocates in a channel equipped with two biased electrodes (see figure 1). The tunnelling current strongly depends on the electronic structure of the bases relative to the electrodes, such as the degree of localization, the geometry and the energy of the electronic states. Nevertheless the tunnelling current measurements allow for a statistical differentiation between the different bases (Lagerqvist et al 2006).
Figure 1. Schematic of sequencing by tunnelling. Reprinted with permission. Copyright 2013 Quantum Biosystems Inc.
Download figure:
Standard image High-resolution imageThe main reason detection using tunnelling currents is potentially orders of magnitude faster than using ionic currents as discussed above is related to the detection of electrical currents with large bandwidths. These can easily reach ranges of over MHz, and possibly tens of MHz for the current amplitudes detected, that is, from tens of pA to nA. For physical reasons, the tunnelling detection scheme is very sensitive to the presence of anything in between the electrodes. When the space between two different bases occupies the gap between electrodes a very small current is detected. However when a particular base occupies the gap between electrodes a substantially higher current flows, which is statistically very distinct from the currents of the other different bases, and is independent of the nearest-neighbour nucleotides (Zwolak and Di Ventra 2005). In addition, the transverse electrical field generated by the electrodes exerts a means of controlling the base dynamics. This helps both the orientation of the bases with respect to the electrodes (Lagerqvist et al 2006), as well as their motion along the channel (He et al 2011), both very important features that help the detection and differentiation of the bases.
The detection of single DNA bases (Tsutsui et al 2010, Chang et al 2010), the differentiation between methylated cytosine and natural cytosine (Tsutsui et al 2011), and finally the sequencing of several DNA oligomers and micro-RNA (Ohshiro et al 2012), have all been recently accomplished using this tunnelling approach. It is indeed impressive that in just a few years since its proposal (Zwolak and Di Ventra 2005), sequencing by tunnelling is approaching the stage of real-life application: a new company—Quantum Biosystems (www.quantumbiosystems.com/)—has been recently founded in Japan precisely to commercialize this particular technology. The developments open up new and exciting possibilities both in the medical field as well as in the study of new phenomena.